
Machine learning of retinal pathology in optical coherence tomography images
Introduction
Of the 7.7 billion world population, approximately 1.3 billion live with some form of vision impairment while 36 million are blind (1,2). Acute macular degeneration (AMD), diabetic retinopathy (DR), central serous retinopathy (CSR) and macular hole (MH) are four common and potentially vision impairing pathologies in the field of ophthalmology. The prevalence of AMD and DR in the United States is estimated to be 1.75 (3) and 4.1 million (4), respectively. Probable US annual incidence for CSR and MH can be inferred from a study limited to Olmsted County, Minnesota which found the annual incidence of CSR and MH to be 9.9 per 100,000 men and 1.7 per 100,000 women (5) and 5.0 per 100,000 men and 11.6 per 1,000,000 women (6), respectively. These four pathologies are often diagnosed using optical coherence tomography (OCT), which is an imaging test that takes cross sectional images of the retina using light waves. However, these four pathologies have some similarities in their presentation on the OCT images which can lead to misdiagnosis.
In the last few years, machine learning and training of deep neural networks has started to emerge in many fields of medicine to help physicians in diagnosis and treatment of patients. For example in the field of dermatology, training of deep neural networks has helped in distinguishing benign from malignant melanoma (7). It has been used to grade brain tumors on histological specimens and magnetic resonance images (8,9). Machine learning has also been used to detect high-grade small bowel obstruction on abdominal radiographs (10) and to detect colorectal polyps on colonoscopy (11).
Positive results have been found in the use of machine learning to diagnose some ophthalmological diseases such as glaucoma (12) and diabetic macular edema (13). The next stage in the field of machine learning is to develop techniques by which the accuracy of the deep neural networks can be improved. One possible method by which this can be done is through data augmentation (DA) and this approach is examined in this analysis. The objective of this research is to examine the effect of DA on the accuracy of a deep neural network on differentiating AMD, CSR, DR, MH and normal OCT images from each other.
Methods
Image gathering
The open source Optical Coherence Tomography Image Database was used to acquire normal OCT images and OCT images for AMD, CSR, DR and MH (14). From this database 55 images of AMD, 102 images of CSR, 107 images of DR, 102 images of MH and 206 normal images were extracted and used in the machine learning analysis. In order to facilitate the machine learning, each set of images was separated into three groups: training, validation and testing. The testing group was fixed at 20 images each, while the training and validation groups accounted for the remaining images in a 70:30 ratio, respectively. The training, validation and testing images were selected randomly from the total images gathered. The images categorized into training, validation and testing were fixed for both the model run with DA and the model run without DA. No preprocessing was performed on any of the images.
Machine learning deep convolutional neural network
Deep convolutional neural networks contain an architecture that is similar to the organization of the human brain. This results in the network being able to process and analyze data in a manner similar to humans (15). Keras (16), an open source neural network library, was used as a deep learning framework in order to retrain the deep convolutional neural network Visual Geometry Group 16 (VGG16) (17). VGG16 was chosen for its high performance level for image classification and its light structure (18-20). VGG16 was developed at Oxford University and contains 13 convolutional layers which process image features ranging from edges and colors to more complex features such as faces. VGG16 also contains 3 fully connected layers which perform nonlinear combination of the image features in the convolutional layers (21). The VGG16 architecture had been pretrained with the ImageNet dataset (22) which contains more than 14 million images. The last three convolutional layers, the last max pooling layer and dense layers of the neural network were retrained in this analysis with the OCT images. The training and validation images were inputted into the model and then afterwards the test images were run in order to analyze how well the model works.
DA
Overfitting is a concern when training neural networks. If the network is not exposed to enough variability in the images or if it is trained on the same images in too many batches it may start to overfit and focus on aspects of the image that are unrelated to the pathology. One method by which overfitting was decreased in this model development was by adding a dropout layer. This resulted in 50% of the neurons to be randomly turned off during the training step resulting in a reduction in the chances of overfitting to occur. Another method by which overfitting may be reduced is through DA.
VGG16 was retrained two times separately, one model with DA and one model without DA. DA consisted of the following changes to the images that were inputted into the model: rotation, width and height shear, horizontal flip and Gaussian noise. Rotation was set to 10% and width and height shear were each set to 10%. This resulted in some of the images that were inputted into VGG16 to be altered based on these parameters before retraining of the model. In addition, Gaussian noise was added to the images before retraining. Gaussian noise consists of distortion of the high frequency features of a model via addition of statistical noise based on the Gaussian distribution. All of these DA techniques help to better account for machine and user variability when taking OCT images.
Overall, two VGG16 models were retrained—one with a dropout layer but without DA and one with both a dropout layer and DA.
Statistical analysis
Statistical analysis of the model’s output on the testing images was performed using R Software [2017] (23). The analysis consisted of sensitivity, specificity, positive predictive value (PPV), negative predictive value (NPV), Matthews correlation coefficient (MCC) and F1 score. MCC quantifies the quality of a binary classification and is often used to assess how well a classification model is performing. It is especially useful when the different categories of images that are being inputted into the model have a different number of images. MCC ranges from –1 to 1 with –1 being a completely inaccurate classifier and 1 being a completely accurate classifier (24). F1 score is another measure of the performance of a classification model. It consists of the harmonic average of the sensitivity and the PPV. F1 score ranges from 0 to 1 and a model with a score closer to 1 is considered to be more accurate.
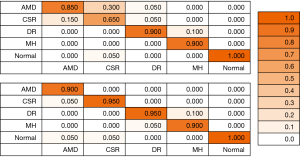
A confusion matrix is created to display the accuracy of the models on the testing images and determine which categories the model had the most confusion with. The columns contain the true classification of the images while the rows represent the model’s prediction on the images.
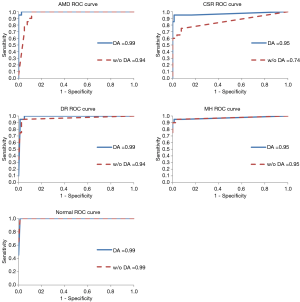
Receiver operating characteristic (ROC) curves are also generated for each of the four ophthalmological pathologies and for normal with and without DA. Area under the curve (AUC) is calculated for each ROC curve. AUC gives insight into how well the model can distinguish between classes with a value closer to 1 indicating a better model.
Results
After the 20 test images for each of the Ophthalmology diseases and for normal were run in the model, sensitivity, specificity, PPV, NPV, MCC and F1 score were calculated as shown in Table 1. This was repeated with the model that used DA. Average sensitivity, specificity, PPV, NPV, MCC and F1 across the five categories of AMD, CSR, DR, MH and normal all increased in value and had smaller standard deviations with the model run with DA as compared to the one without. Specifically, MCC, which is often used as a benchmark to measure how well a model works had an average increase from 0.83 to 0.93.
Table 1
| Ophthalmological condition | Sensitivity | Specificity | PPV | NPV | MCC | F1 Score |
|---|---|---|---|---|---|---|
| Without data augmentation | ||||||
| AMD | 0.850 | 0.913 | 0.708 | 0.961 | 0.714 | 0.773 |
| CSR | 0.650 | 0.950 | 0.765 | 0.916 | 0.639 | 0.703 |
| DR | 0.900 | 0.975 | 0.900 | 0.975 | 0.875 | 0.900 |
| MH | 0.900 | 1.000 | 1.000 | 0.976 | 0.937 | 0.947 |
| Normal | 1.000 | 0.988 | 0.952 | 1.000 | 0.970 | 0.976 |
| Average ± SD | 0.860±0.116 | 0.965±0.031 | 0.865±0.111 | 0.965±0.028 | 0.827±0.129 | 0.860±0.105 |
| With data augmentation | ||||||
| AMD | 0.900 | 1.000 | 1.000 | 0.976 | 0.937 | 0.947 |
| CSR | 0.950 | 0.988 | 0.950 | 0.988 | 0.938 | 0.950 |
| DR | 0.950 | 0.975 | 0.905 | 0.987 | 0.908 | 0.927 |
| MH | 0.900 | 0.988 | 0.947 | 0.975 | 0.905 | 0.923 |
| Normal | 1.000 | 0.975 | 0.909 | 1.000 | 0.941 | 0.952 |
| Average ± SD | 0.940±0.037 | 0.985±0.009 | 0.942±0.034 | 0.985±0.009 | 0.926±0.016 | 0.940±0.005 |
PPV, positive predictive value; NPV, negative predictive value; MCC, Matthews correlation coefficient; AMD, Acute macular degeneration; CSR, central serous retinopathy, DR, diabetic retinopathy; MH, macular hole.
A confusion matrix, shown in Figure 1, was also created in order to better understand which of the categories of AMD, CSR, DR, MH and normal the model had the most confusion with. This was again performed for both the model with and without DA. Each of the Ophthalmology diseases and the normal OCT group stayed the same or increased in accuracy for the test images once DA was added to the model.
Further, receiver operating curves (ROC) were developed for each of the Ophthalmology diseases and for normal OCT and area under the curve (AUC) was calculated for each ROC. Figure 2 shows these ROC and the AUC for the models run with and without DA. The average AUC increased from 0.91 without DA to 0.97 with DA.
Discussion
The confusion matrix shows that the model without DA had significant difficulty in distinguishing AMD from CSR. Of the true CSR test images the model predicted 65% of them to be CSR and 30% to be AMD. Further, of the true AMD test images the model predicted 85%, second lowest of the five categories, to be AMD and the remaining 15% to be CSR. In the model run with DA of the true CSR test images, 95% were predicted to be CSR and of the true AMD test images, 90% were predicted to be AMD, indicating that DA significantly helped the model to better distinguish CSR from AMD and vice versa. It is possible that AMD and CSR had increased confusion because of the limited number of total images of AMD that were inputted into the model as compared to the other diseases and normal. Further CSR has some similar qualities on OCT as AMD and can be hard to distinguish (25). Another indicator that the number of images may have played a factor in the lower accuracy for some diseases than others is that the category of normal OCT had the highest number of images inputted into the model and without DA the model had 100% test accuracy. As such, the DA techniques used in this analysis may be especially useful when there are a limited number of training images.
The AUC for each of the ROC curves increased for the model run with DA as compared to the one without. AUC for CSR had the highest increase from 0.74 to 0.95. In addition, AUC for AMD jumped from third highest (0.94) in the model without DA to the highest (0.99) in the model with DA. These results again indicate the value of the DA techniques when the images are limited and/or when the images have similar features that may make it harder to distinguish from one another.
Other research
Research has been performed on machine learning for various ophthalmological pathologies using OCT images (26-28) and multispectral microscopy images (29). Even though OCT machines have some standardization and set calibration, there exists inter-machine variability and user generated variability. This variability combined with the variability in the presentation of each ophthalmological pathology makes it very difficult for deep neural networks to improve in their accuracy significantly. DA can help to generate variability from one image so that the model is able to analyze this variability during the training stage rather than seeing it for the first time during the testing stage. Since neural networks are often trained using data obtained from one machine in one location, DA can help to account for some of the variability between machines and user generated variability. Further, DA can be especially useful when there are a limited number of images available for input into the model. For example, in this analysis, only a total of 55 AMD images were gathered from the database. After setting aside images for testing, the deep convolutional neural network was limited to a relatively small number of images for training and validation. This may have led to some of the confusion between AMD and CSR. However, after DA the model seemed to be better able to distinguish AMD from other pathologies as indicated by the increase in statistical measures such as MCC and in AUC.
Future
The model was trained on a limited number of OCT images. Acquiring a larger quantity and better quality OCT images can help to improve the accuracy of the model. Next steps in this field of research include creating an application in which physicians can input an OCT image and the top three diagnoses with percentages are outputted. This can be used by physicians as a tool to strengthen support for their own diagnosis or to consider a diagnosis that was low on their differential but high on the model’s. Further, OCT is an imaging technique used not only by ophthalmologists but also by cardiologists in imaging coronary arteries, by oncologists for detecting esophageal dysplasia and by dermatologists for detecting skin carcinomas. As such it may be possible for non ophthalmological medical professionals to use deep convolutional neural networks as an aid in detecting pathologies in OCT images in their field of medicine.
Limitations
The current model in this analysis has only been trained to identify AMD, CSR, DR, MH and normal OCT images. It will have to be properly trained to decipher other OCT image pathologies. The model does not account for the patient’s symptoms or time course of the symptoms which can significantly influence the differential diagnosis. Further, automated classification of images is very difficult because of the variability in the image (brightness, zoom, rotation etc) and because of the variability in the presentation of the pathology in the image.
Conclusions
The deep neural network trained was able to distinguish AMD, CSR, DR and MH and normal eye OCT images with appreciable MCC and F1 score. These and other statistical measures of the model increased when the same model was run with DA techniques, including rotation, shear, flipping and Gaussian noise. Further, the AUC for the ROC curves for each of the ophthalmological diseases and for normal increased once DA was added. The average AUC for the model using DA increased by 0.06 to 0.97, indicating that model performs well. The model without DA had the greatest difficulty in distinguishing AMD from CSR. The addition of DA significantly reduced this confusion. Future steps in this field include obtaining larger numbers and higher quality of OCT images in order to further improve the model. Further, creating an application with this model can serve as an aid for physicians in real-time diagnosing of OCT images.
Acknowledgments
Funding: None.
Footnote
Conflicts of Interest: The author has completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/jmai.2019.08.01). The author has no conflicts of interest to declare.
Ethical Statement: The author is accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Bourne RRA, Flaxman SR, Braithwaite T, et al. Magnitude, temporal trends, and projections of the global prevalence of blindness and distance and near vision impairment: a systematic review and meta-analysis. Lancet Glob Health 2017;5:e888-97. [Crossref] [PubMed]
- WHO. Blindness and vision impairment. Accessed June 10, 2019. Available online: https://www.who.int/news-room/fact-sheets/detail/blindness-and-visual-impairment
- Friedman DS, O'Colmain BJ, Muñoz B, et al. Prevalence of age-related macular degeneration in the United States. Arch Ophthalmol 2004;122:564-72. [Crossref] [PubMed]
- Kempen JH, O'Colmain BJ, Leske MC, et al. The prevalence of diabetic retinopathy among adults in the United States. Arch Ophthalmol 2004;122:552-63. [Crossref] [PubMed]
- Kitzmann AS, Pulido JS, Diehl NN, et al. The incidence of central serous chorioretinopathy in Olmsted County, Minnesota, 1980-2002. Ophthalmology 2008;115:169-73. [Crossref] [PubMed]
- McCannel CA, Ensminger JL, Diehl NN, et al. Population-based incidence of macular holes. Ophthalmology 2009;116:1366-9. [Crossref] [PubMed]
- Esteva A, Kuprel B, Novoa RA, et al. Dermatologist-level classification of skin cancer with deep neural networks. Nature 2017;542:115-8. [Crossref] [PubMed]
- Ker J, Bai Y, Lee HY, et al. Automated brain histology classification using machine learning. J Clin Neurosci 2019;66:239-45. [Crossref] [PubMed]
- Banzato T, Causin F, Della Puppa A, et al. Accuracy of Deep Learning to Differentiate the Histopathological Grading of Meningiomas on MR Images: A Preliminary Study. J Magn Reson Imaging 2019; [Epub ahead of print]. [Crossref] [PubMed]
- Cheng PM, Tejura TK, Tran KN, et al. Detection of high-grade small bowel obstruction on conventional radiography with convolutional neural networks. Abdom Radiol (NY) 2018;43:1120-7. [Crossref] [PubMed]
- Mori Y. Kudo Se, Misawa M. Detecting colorectal polyps with use of artificial intelligence. J Med Artif Intell 2019;2:11. [Crossref]
- Ahn JM, Kim S, Ahn KS, et al. A deep learning model for the detection of both advanced and early glaucoma using fundus photography. PLoS One 2018;13:e0207982. [Crossref] [PubMed]
- Kamble RM, Chan GCY, Perdomo O, et al. Automated Diabetic Macular Edema (DME) Analysis Using Fine Tuning with Inception-Resnet-v2 on OCT Images. Conf Proc IEEE Eng Med Biol Soc 2018;2018:2715-8.
- Gholami P, Roy P, Parthasarathy MK, et al. OCTID: Optical Coherence Tomography Image Database. 2019. arXiv:1812.07056.
- Yates EJ, Yates LC, Harvey H. Machine learning "red dot": open-source, cloud, deep convolutional neural networks in chest radiograph binary normality classification. Clin Radiol 2018;73:827-31. [Crossref] [PubMed]
- Keras: The Python Deep Learning library. Available online: https://keras.io
- Simonyan K, Zisserman A. Very Deep Convolutional Networks for Large-Scale Image Recognition. 2015. arXiv:1409.1556.
- Santin M, Brama C, Théro H, et al. Detecting abnormal thyroid cartilages on CT using deep learning. Diagn Interv Imaging 2019;100:251-7. [Crossref] [PubMed]
- He K, Zhang X, Ren S, et al. Deep Residual Learning for Image Recognition. In: 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR). 2016. doi:
10.1109/CVPR.2016.90 . - Szegedy C, Vanhoucke V, Ioffe S, et al. Rethinking the Inception Architecture for Computer Vision. In: 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR). Las Vegas, NV, USA: 2016. doi:
10.1109/CVPR.2016.308 . - Orita K, Sawada K, Koyama R, Ikegaya Y. Deep learning-based quality control of cultured human-induced pluripotent stem cell-derived cardiomyocytes. J Pharmacol Sci 2019; [Epub ahead of print]. [Crossref] [PubMed]
- Russakovsky O, Deng J, Su H, et al. ImageNet Large Scale Visual Recognition Challenge. 2014. arXiv:1409.0575.
-
The R Project for Statistical Computing - Aggarwal SLP. Data augmentation in dermatology image recognition using machine learning. Skin Res Technol 2019; [Epub ahead of print]. [Crossref] [PubMed]
- Ahn SJ, Kim TW, Huh JW, et al. Comparison of features on SD-OCT between acute central serous chorioretinopathy and exudative age-related macular degeneration. Ophthalmic Surg Lasers Imaging 2012;43:374-82. [Crossref] [PubMed]
- Sun Z, Sun Y. Automatic detection of retinal regions using fully convolutional networks for diagnosis of abnormal maculae in optical coherence tomography images. J Biomed Opt 2019;24:1-9. [PubMed]
- Fu H, Baskaran M, Xu Y, et al. A Deep Learning System for Automated Angle-Closure Detection in Anterior Segment Optical Coherence Tomography Images. Am J Ophthalmol 2019;203:37-45. [Crossref] [PubMed]
- Schmidt-Erfurth U, Sadeghipour A, Gerendas BS, Waldstein SM, Bogunović H. Artificial intelligence in retina. Prog Retin Eye Res 2018;67:1-29. [Crossref] [PubMed]
- Habibalahi A, Bala C, Allende A, et al. Novel automated non invasive detection of ocular surface squamous neoplasia using multispectral autofluorescence imaging. Ocul Surf 2019;17:540-50. [Crossref] [PubMed]
Cite this article as: Aggarwal P. Machine learning of retinal pathology in optical coherence tomography images. J Med Artif Intell 2019;2:20.


