Deep learning applications in coronary anatomy imaging: a systematic review and meta-analysis
Highlight box
Key findings
• Deep learning has important applications in coronary anatomy imaging.
• CT-FFR is an example which has translated into clinical practice and patients’ care.
• CNNs have been the most powerful in recent literature.
What is known and what is new?
• Coronary anatomy imaging is mainly assessed by human experts.
• Deep learning has shown a high performance in coronary anatomy interpretation, prediction, and improving patient care and safety.
What is the implication, and what should change now?
• Research in deep learning for coronary anatomy imaging is making significant advancements.
• Successful deep learning applications will require clinical validation.
Introduction
Background
Coronary artery disease (CAD) is considered a leading cause of death and hospitalisation in high-income countries, and worldwide (1). The progressive nature of coronary atherosclerosis is the main underlying pathological process. Therefore, it is essential to have timely diagnosis of CAD to aid the management of patients and reduce both morbidity and mortality.
The last two decades have witnessed significant advancements in CAD imaging, from functional assessment of coronary artery stenoses and how they impact on the myocardium at stress and rest, using cardiac magnetic resonance (CMR), myocardial perfusion scintigraphy (MPS), and echocardiography, to anatomical assessment by means of coronary computed tomography angiography (CCTA) and invasive X-rays coronary angiography.
Computer vision technology on the other hand is going through an exciting era following the revolution of deep learning and artificial intelligence (AI) algorithms. CAD imaging is one of the key applications which has been targeted by many computer vision experts and deep learning practitioners.
Rationale and objectives
There has been an explosion in the number of deep learning publications in CAD over the recent years with a focus on atherosclerosis and coronary anatomy imaging. The wide range of methodology presented in the recent literature opened the door for applications in various coronary artery imaging modalities.
The mounting volume of new literature has left clinicians with a two-fold challenge: first of how to deal with increasing volume of new information on CAD diagnosis, prognosis, and risk stratification, and second of how far can we trust the evidence of machine learning and deep learning algorithms to make decisions on patients’ care.
This review aims to unravel this challenge by summarising the new information we gained so far in this field, evaluating the performance of the presented deep learning algorithms, and drawing some conclusions on potential meaningful applications. We present the following article in accordance with the PRISMA reporting checklist (available at https://jmai.amegroups.com/article/view/10.21037/jmai-22-36/rc).
Methods
Design
This review follows the Cochrane Review structure of diagnostic test accuracy (DTA) (2). The umbrella protocol for this systematic review is registered in the International Prospective Register of Systematic Reviews (PROSPERO, CRD42020204164), and reported according to PRISMA guidelines. All searching activities were performed by two independent reviewers (EA and UD), with divergences solved after consensus.
The PICO approach was used to define the main review question:
- Population: adults’ cohort with suspected or known CAD;
- Intervention: deep learning applications in coronary atherosclerosis imaging;
- Comparison: comparison with conventional coronary atherosclerosis imaging;
- Outcome: improve test accuracy and patient care.
Selection criteria
Without restrictions on minimal sample sizes or recruitment process, both prospective and retrospective studies were included. The included studies had participants with known or suspected CAD who had atherosclerosis imaging (invasive and non-invasive) with the application of deep learning technology, and compared with the gold standard (reference) test used in clinical practice.
Competitions presented in conferences on deep learning techniques, such as at the Medical Image Computing and Computer Assisted Intervention (MICCAI) conference, animal studies, and simulation studies were not included due to ambiguity in their direct relation to patient care. Studies which used atherosclerosis data as a target for outcome prediction were excluded, as were studies, which focused on clinical data and imaging reports rather than imaging data for prediction. Studies, which used deep learning software with no details on the deep learning architecture were also excluded. Fusion imaging studies were not part of this review, and studies of automated coronary anatomy and atherosclerosis quantification, which relied mainly on hand crafted or non-learning algorithms were not included.
For fractional flow reserve (FFR) derived from CCTA using deep learning, only the original publications were included in this review, all subsequent publications, which used the same algorithms for different clinical applications were considered external validation papers and were not included in this review.
Search procedure
MEDLINE (with PubMed extension) and EMBASE using Ovid search engine was conducted to search the published literature. Yale Mesh Analyzer was used to include all possible Medline Subject Headings (MeSH) terms, after identifying two studies manually on MEDLINE database with focus on deep learning and CAD atherosclerosis imaging modalities. The PMIDs for those papers were extracted and inserted into the analyser, these produced Mesh terms to guide the systematic search. Truncation has been used in imaging term: [‘coronar*’], [‘myocardia*’], [‘atherosclero*’], [‘isch?mi*’], and [‘calci*’]. Plain terms were used for [‘machine learning’], [‘deep learning’], [‘artificial intelligence’], [‘neural networks’], [‘unsupervised learning’], [‘supervised learning’], [‘semi-supervised learning’], [‘heart’], [‘plaque’], and [‘stenosis’]. The search included all records from database inception until 21st of October 2020 with no language constraints. Data was collected by EA and UD. Full Ovid search strategy and output is shown in https://cdn.amegroups.cn/static/public/jmai-22-36-1.pdf. Due to reports of missing relevant studies and inconsistency using methodology search filters (2), this approach has not been used.
Search results
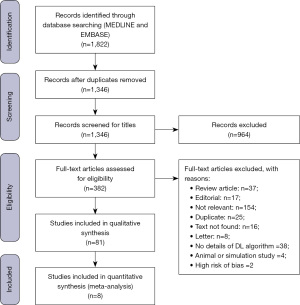
Search results yielded 81 studies to be used for the systematic review and only a subset of 8 studies with unified defined outcomes were used for meta-analysis. Search results are shown in Figure 1.
Data extraction
The summary of input data, which were extracted from each study are reported below:
- First author’s surname;
- Year of publication;
- Total number of participants (images if not available);
- Imaging modality used for deep learning;
- Index test;
- Reference test;
- Deep learning techniques;
- External validation;
- Model performance metrics.
Assessment of risk of bias
The Quality Assessment of Diagnostic Accuracy Studies (QUADAS) tool was used to assess the risk of bias. Five main fields were assessed using a modified version:
- Patient selection: randomly selected patients from a population meeting the inclusion criteria is considered a high-quality study;
- Index test: including a comparator test is expected in a high-quality diagnostic test study;
- Reference test: a gold standard test for validation is mandatory in all high-quality diagnostic test studies;
- Index test results blinded: the results of the comparator test are expected to be blinded to the deep learning arm in a high-quality study;
- Reference test results blinded: the results of the gold standard test are expected to be blinded to the deep learning arm in a high-quality study.
Statistical analysis
The performance of deep learning models was measured with various metrics including sensitivity, specificity, area under the curve (AUC), precision, recall, F1 score, Dice coefficient, Jaccard coefficient, and correlation. Those metrics were described quantitatively.
Data were reported as count or percentages. The pooled values of some of the reported diagnostic accuracy after the application of deep learning models, which were part of the meta-analysis, were visualised by forest plots.
A confusion matrix was produced for each of the included studies in meta-analysis given that most studies did not report the true negative (TN), true positive (TP), false negative (FN), and false positive (FP) values. This was calculated by taking sample size (S) to calculate FN from sensitivity, and FP from specificity. The TN and TP were then calculated from total sample size S.
Meta-analysis was performed on studies, which reported the same outputs with the corresponding sensitivity and specificity. Since pooling sensitivities or specificities can be misleading, the diagnostic odds ratio (DOR) approach is taken to calculate the pooled diagnostic performance. The fixed effect case of Mantel-Haenszel (MH) method is used.
Heterogeneity was examined using tau2, I2 and Q tests. P value of less than 0.05 was considered statistically significant.
All statistical analysis was performed using RStudio software version 1.4.1106 using R 4.0.4 programming language.
Results
Characteristics of studies
The final number of studies included in this systematic review was 81, all published over 6 years between 2015 and 2020, which indicates the recency of this topic.
Details of first author, year of publication, sample size, deep learning and machine learning techniques, index test (comparator) and reference test (gold standard) are shown in Table 1.
Table 1
| First author | Year | Model output | Sample size | Imaging modality | Model | Index test | Reference test | External validation |
|---|---|---|---|---|---|---|---|---|
| Rodrigues et al. (3) | 2016 | Pericardial and mediastinal fat classification | 20 | CCTA | RF | Manual feature extraction algorithms | Expert reader | No |
| Kang et al. (4) | 2015 | Coronary stenosis classification | 42 | CCTA | SVM | Expert reader | Invasive coronary angiography | No |
| Araki et al. (5) | 2016 | Coronary plaque calcification | 15 | IVUS | SVM | NA | cIMT | No |
| Itu et al. (6) | 2016 | FFR prediction | 87 | CCTA | MLP | Computational fluid dynamics CT-FFR | Invasive FFR | Yes |
| Wolterink et al. (7) | 2016 | CAC quantification | 250 | CCTA | CNN | NA | Expert reader | No |
| Su et al. (8) | 2017 | Media adventitia border detection | 4 | IVUS | MLP | NA | Expert reader | No |
| Yong et al. (9) | 2017 | Coronary lumen segmentation | 64 | OCT | CNN | NA | Expert reader | No |
| Xu et al. (10) | 2017 | Coronary plaque classification | 18 | OCT | CNN and SVM | NA | Expert reader | No |
| Zreik et al. (11) | 2019 | Coronary plaque classification | 163 | CCTA | RNN | NA | Expert reader | No |
| Zreik et al. (12) | 2018 | LV segmentation for coronary stenosis significance classification | 156 | CCTA | CNN + SVM | NA | Invasive FFR | No |
| Kolluru et al. (13) | 2018 | Coronary plaque classification | 48 | OCT | CNN | NA | Expert reader | No |
| Zhang et al. (14) | 2018 | Coronary plaque classification | 61 | IVUS | SVM | NA | Expert reader | No |
| Oh et al. (15) | 2018 | Lipid core plaque detection | 116 | IVUS | CNN | NA | Expert reader | No |
| van Rosendael et al. (16) | 2018 | Clinical outcome prediction | 8,844 | CCTA | Boosted ensemble algorithm | Conventional clinical risk scores | Clinical outcomes | No |
| Stuckey et al. (17) | 2018 | CAD detection | 606 | cPSTA | Elastic net | NA | Invasive coronary angiography | No |
| Lessmann et al. (18) | 2018 | CAC detection | 1,744 | CCTA | CNN | NA | Expert reader | No |
| Šprem et al. (19) | 2018 | Motion artefact detection in CACS | 585 | CCTA | CNN | NA | Conventional CACS | No |
| Hae et al. (20) | 2018 | Prediction of myocardium subtended by coronary stenosis | 932 | CCTA | SVM | NA | Invasive coronary angiography | Yes |
| Dey et al. (21) | 2018 | FFR prediction | 254 | CCTA | Boosted ensemble algorithm | Conventional CCTA | Invasive FFR | No |
| van Hamersvelt et al. (22) | 2019 | LV segmentation for coronary stenosis significance classification | 126 | CCTA | SVM | NA | Expert reader | No |
| Cho et al. (23) | 2019 | FFR classification | 1,501 | Invasive coronary angiography | XGBoost | NA | Invasive FFR | Yes |
| Liu et al. (24) | 2019 | Vulnerable plaque detection | 2,300 (images) | OCT | CNN | NA | Expert reader | No |
| Gessert et al. (25) | 2019 | Coronary plaque segmentation | 49 | OCT | CNN | NA | Expert reader | No |
| Abdolmanafi et al. (26) | 2019 | Coronary artery wall pathology detection | 45 | OCT | CNN | NA | Expert reader | No |
| Liu et al. (27) | 2019 | Bifurcation lesion detection | 308 | Invasive coronary angiography | CNN | NA | Expert reader | No |
| Gharaibeh et al. (28) | 2019 | CAC quantification | 34 | IVUS | CNN | NA | Expert reader | No |
| Jun et al. (29) | 2019 | Thin cap fibroatheroma classification | 100 | IVUS | CNN | NA | OCT | No |
| Lee et al. (30) | 2019 | Coronary artery segmentation | 4,980 | Invasive coronary angiography | CNN | NA | Expert reader | No |
| Yang et al. (31) | 2019 | Coronary artery segmentation | 2,042 | Invasive coronary angiography | CNN | NA | Expert reader | Yes |
| Wang et al. (32) | 2019 | Media adventitia border detection | 22 | IVUS | MLP | P6 and P8 detectors | Expert reader | No |
| Johnson et al. (33) | 2019 | Clinical outcome prediction | 6,892 | CCTA | KNN | Conventional CT and clinical risk scores | Clinical outcomes | No |
| Kolossváry et al. (34) | 2019 | Coronary plaque classification | 21 | CCTA | Least angle regression + radiomics | Histogram assessment by expert reader | Histology (ex vivo) | No |
| Wang et al. (35) | 2019 | FFR prediction | 63 | CCTA | RNN | Conventional CCTA | Invasive FFR | No |
| Datong et al. (36) | 2019 | CAC detection | 820 (images) | CCTA | CNN | NA | Expert reader | No |
| Oikonomou et al. (37) | 2019 | Clinical outcome prediction | 5,487 | CCTA | RF + radiomics | Conventional clinical risk scores | Clinical outcomes | Yes |
| Masuda et al. (38) | 2019 | Coronary plaque classification | 78 | CCTA | Extreme gradient boosting | Conventional CCTA | IVUS | No |
| Kigka et al. (39) | 2019 | Coronary plaque progression prediction | 40 | CCTA | RF | NA | Clinical outcomes | No |
| Zhang et al. (40) | 2019 | Coronary risk prediction | 4,415 | CCTA | Boosted ensemble algorithm | Conventional clinical risk scores | Clinical outcomes | No |
| Commandeur et al. (41) | 2019 | Epicardial adipose tissue quantification | 850 | CCTA | CNN | NA | Expert reader | No |
| Hong et al. (42) | 2019 | Coronary artery segmentation | 156 | CCTA | CNN | NA | Expert reader | No |
| Huo et al. (43) | 2019 | CAC detection | 2,332 | CCTA | CNN | NA | Expert reader | No |
| Wang et al. (44) | 2020 | MPVI prediction | 9 | IVUS | SVM and RF | GLMM | Follow-up MPVI | No |
| Lee et al. (45) | 2020 | FFR prediction | 1,328 | IVUS | AdaBoost | NA | Invasive FFR | No |
| Wu et al. (46) | 2020 | Coronary stenosis detection | 63 | Invasive coronary angiography | CNN | NA | Expert reader | No |
| Sampedro-Gómez et al. (47) | 2020 | Stent restenosis prediction | 263 | Invasive coronary angiography | ERT | Conventional clinical risk scores | Clinical outcomes | No |
| Miyoshi et al. (48) | 2020 | Coronary neointimal coverage classification, yellow colour classification, red thrombus detection | 107 | Invasive coronary angioscopy | GAN | SVM | Expert reader | Yes |
| Zhang et al. (49) | 2020 | Coronary stenosis classification | 228 | Invasive coronary angiography | HEAL | NA | Expert reader | No |
| Du et al. (50) | 2021 | Coronary artery segmentation, stenosis classification, total occlusion detection, calcification detection, thrombus detection, dissection detection | 10,073 | Invasive coronary angiography | CNN and GAN | NA | Expert reader | No |
| He et al. (51) | 2020 | Coronary plaque segmentation | 24 | OCT | CNN | NA | Expert reader | No |
| Yabushita et al. (52) | 2021 | Coronary artery segmentation | 146 | Invasive coronary angiography | CNN | NA | Expert reader | No |
| Hamaya et al. (53) | 2020 | Clustering epicardial functional stenosis with low CFR | 364 | Invasive coronary angiography | Unsupervised hierarchical clustering | K-mean clustering | Clinical outcomes | No |
| Lee et al. (54) | 2019 | Coronary plaque segmentation | 55 | OCT | CNN | A-line CNN detector | Expert reader | No |
| Min et al. (55) | 2020 | Thin cap fibroatheroma classification | 602 | OCT | CNN | NA | Expert reader | No |
| Commandeur et al. (56) | 2020 | Clinical outcome prediction | 1,912 | CCTA | Extreme gradient boosting | Conventional CT and clinical risk scores | Clinical outcomes | No |
| Muscogiuri et al. (57) | 2020 | CAD classification | 288 | CCTA | CNN | NA | Expert reader | No |
| Benz et al. (58) | 2020 | Coronary artery image reconstruction | 43 | CCTA | CNN | Adaptive statistical iterative reconstruction | Invasive coronary angiography | No |
| Wang et al. (59) | 2020 | CAC quantification | 530 | CCTA | CNN | NA | Expert reader | No |
| Al’Aref et al. (60) | 2020 | Coronary stenosis prediction from CACS | 13,054 | CCT | Boosted ensemble algorithm | NA | CCTA | No |
| Kawasaki et al. (61) | 2020 | FFR prediction | 47 | CCTA | RF | NA | Invasive FFR | No |
| Fischer et al. (62) | 2020 | CAC quantification | 200 | CCTA | RNN | NA | Expert reader | No |
| van Velzen et al. (63) | 2020 | CAC quantification | 7,240 | CCTA | CNN | NA | Expert reader | No |
| Zreik et al. (64) | 2020 | Coronary stenosis classification | 187 | CCTA | CNN + SVM | NA | Invasive FFR | No |
| Kumamaru et al. (65) | 2020 | FFR prediction | 1,052 | CCTA | CNN + GAN | Conventional CCTA | Invasive FFR | No |
| Candemir et al. (66) | 2020 | Coronary stenosis classification | 493 | CCTA | CNN | NA | Expert reader | Yes |
| Shu et al. (67) | 2022 | Clinical outcome prediction | 154 | CCTA | SVM + radiomics | NA | Expert reader | Yes |
| van den Oever et al. (68) | 2020 | CAC rule out | 100 | CCTA | CNN | NA | Expert reader | Yes |
| Han et al. (69) | 2020 | Coronary stenosis classification | 150 | CCTA | CNN | Expert reader | Invasive coronary angiography | No |
| Han et al. (70) | 2020 | Rapid plaque progression prediction | 1,083 | CCTA | Boosted ensemble algorithm | Conventional clinical risk scores | Clinical outcomes | No |
| Lin et al. (71) | 2020 | Pericoronary adipose tissue prognosis prediction | 177 | CCTA | Boosted ensemble algorithm + radiomics | Conventional CT and clinical risk scores | Clinical outcomes | No |
| Chen et al. (72) | 2020 | Coronary artery segmentation | 124 | CCTA | CNN | Expert reader | Invasive coronary angiography | No |
| Tesche et al. (73) | 2021 | Clinical outcome prediction | 361 | CCTA | Boosted ensemble algorithm | Conventional CT and clinical risk scores | Clinical outcomes | No |
| Al’Aref et al. (74) | 2020 | CL precursors detection | 46 | CCTA | Boosted ensemble algorithm | Traditional CCTA CL precursors | Invasive coronary angiography | Yes |
| Hong et al. (75) | 2020 | CCTA image noise reduction | 82 | CCTA | CNN | NA | Invasive coronary angiography | No |
| Podgorsak et al. (76) | 2020 | Coronary segmentation and FFR prediction | 64 | CCTA | CNN | Expert reader | Invasive FFR | No |
| Eberhard et al. (77) | 2020 | FFR prediction | 56 | CCTA | CNN | Invasive FFR | Clinical outcomes | No |
| Son et al. (78) | 2020 | CAC prediction | 20,130 | Retinal fundus imaging | CNN | NA | CCTA | No |
| Carson et al. (79) | 2020 | FFR prediction | 25 | CCTA | MLP and RNN | MPR | Invasive FFR | Yes |
| Gangl et al. (80) | 2019 | Coronary plaque segmentation | 104 (images) | OCT | CNN | NA | Expert reader | No |
| Głowacki et al. (81) | 2020 | Coronary stenosis prediction from CACS | 435 | CCT | Extreme gradient boosting | NA | CCTA | No |
| Hoshino et al. (82) | 2020 | FAI clusters | 220 | CCTA | Unsupervised hierarchical clustering | Invasive FFR | Clinical outcomes | No |
| Kawaguchi et al. (83) | 2018 | FFR prediction | 934 | CCTA | CNN | NA | Invasive FFR | No |
CCTA, coronary computed tomographic angiography; RF, random forest; SVM, support vector machine; IVUS, intra-vascular ultrasound; NA, not available; cIMT, carotid intima-media thickness; FFR, fractional flow reserve; MLP, multi-layer perceptron; CT, computed tomography; CAC, coronary artery calcification; CNN, convolutional neural network; OCT, optical coherence tomography; RNN, recurrent neural network; LV, left ventricle; CAD, coronary artery disease; cPSTA, cardiac phase space tomography analysis; CACS, coronary artery calcium score; KNN, k-nearest neighbours; MPVI, morphological plaque vulnerability index; GLMM, generalised linear mixed model; ERT, extremely randomised tree; GAN, generative adversarial network; HEAL, hierarchical attentive multi-view; CFR, coronary flow reserve; CL, culprit lesion; MPR, multi-variant polynomial regression; FAI, fat attenuation index.
The most popular imaging modality in deep learning application was CCTA (58%), as shown in Figure 2. However, invasive coronary angiography has gained more interest in recent years, along with invasive coronary intra-vascular imaging [optical coherence tomography (OCT) and intravascular ultrasound (IVUS)], which have been a focus for deep learning applications in recent years. Both OCT and IVUS are performed during invasive coronary angiography to add more detailed imaging analysis of atherosclerotic lesions seen on Cine X-ray images.
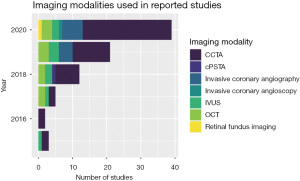
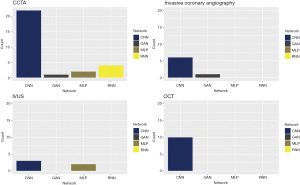
The most commonly used deep learning technique was convolutional neural network (CNN) as shown in Figure 3, with more than half of the studies (52%) have used this approach as a single model or combined with other models. The use of multi-layer perceptron (MLP) was scarce with only 4 studies reported their results using MLP approach. There was a variety of models used with only a few studies in each category, including generative adversarial network (GAN), recurrent neural network (RNN), random forest (RF), gradient boost, support vector machine (SVM), to name a few.
Principle deep learning applications and meta-analysis
Coronary calcification
Several CCTA studies have focused on detection or quantification of coronary calcium given its prognostic importance in clinical outcomes. There have been successful applications of deep learning models using mainly CNNs to detect coronary artery calcification (CAC). Studies with large sample sizes have been conducted and reported good or excellent model performance in detecting CAC. Huo et al. (43) used 2,332 of scan-rescan pairs as input to their CNN architecture called AID-Net, which is composed of 3D ResNet and 3D DenseNet layers. They reported high model performance with AUC as high as 0.93 in detecting CAC. van Velzen et al. (63) used a large sample of CCTA data from 7,240 participants, and with a CNN they quantified CAC and achieved a high model performance with 97% inter-class correlation with expert reader and 96% accuracy. All other studies had smaller sample sizes and reported similar level of performance for CAC detection and quantification using CNNs.
Fischer et al. (62) used RNN for CAC quantification, and their model achieved good performance with sensitivity of 92% and specificity of 89%. All these reports confirm that deep learning algorithms are capable of performing CAC detection or rule out, and quantification in a highly reliable way and with less time than an expert human reader.
Coronary artery stenosis
All of the four main imaging modalities (CCTA, OCT, IVUS, invasive coronary angiography) were used for deep learning applications to assess coronary stenosis in various ways: coronary plaque classification and segmentation, coronary stenosis classification and segmentation, culprit lesions predictors, vulnerable plaque precursors, thrombus, dissection and clinical outcome prediction.
Invasive coronary angiography studies used large numbers of patients for coronary artery segmentation. Du et al. (50) looked at 10,073 cases and trained a CNN and a GAN for better characterisation of coronary lesion location and description. Their model was able to perform coronary artery segmentation, stenosis classification, detection of total occlusion, calcification, thrombus and coronary dissection. They reported an AUC of 0.8 for coronary stenosis classification and F1 score of 0.82, and similar metrics for the other outputs were achieved, with a better performance in coronary segmentation with an AUC of 0.86. Similar performance was achieved from CCTA studies in coronary artery segmentation, Chen et al. (72) reported an AUC of 0.89 after using a CNN with 3D U-Net architecture on a sample size of 432 cases.
FFR
The earliest and most successful application of deep learning in atherosclerosis and coronary anatomy imaging was achieved in the assessment of FFR using CCTA, currently there are clinical applications available and it has gained a lot of attention in cardiovascular medicine and cardiothoracic surgery, due to the advantage of assessing coronary anatomy and ischaemic burden of coronary lesions both non-invasively.
The first application was in 2016 when Itu et al. (6) analysed 87 cases of CCTA and used a MLP architecture and some feature extraction techniques to calculate reliable FFR values, which was validated by invasive measurements. Also, this was compared to conventional CT-FFR based on computational fluid dynamics and showed to be more efficient. Their reported specificity was 84% and sensitivity 82% compared to invasive assessment. Many studies have been published since then to externally validate those findings and the algorithm has been tested for various applications beyond just the absolute FFR values, such as looking at clinical outcome and prognosis.
Following this successful application, several studies have used more developed deep learning techniques to predict CT-FFR using CNNs and RNNs, and they all reported high performance metrics after comparing with invasive FFR.
A meta-analysis has been performed on eight studies which reported FFR prediction and had sensitivity and specificity reported. Figure 4 shows a coupled forest plot for sensitivity and specificity to assess heterogeneity by visual appreciation.

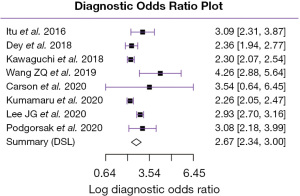
After calculating the DOR for all studies using MH method, the pooled value of DOR was estimated at 12.5. According to the literature this is considered as a positive finding as it is higher than 10 (84). Figure 5 shows a forest plot of the natural logarithmic DOR (lnDOR) for all eight studies with the pooled value in the summary (MH).
Assessment of heterogeneity
Quantifying heterogeneity of the eight studies included in meta-analysis showed tau2 =0.0011 with confidence interval (0.0000, 0.0166), this indicates no significant heterogeneity between studies.
I2 was calculated at 22.6%, indicating that true effect size differences have affected less than quarter of the variation in our data. According to “rule of thumb” from the literature, heterogeneity based on this value is considered mild.
The predictive interval was ranging from (0.9006 to 1.1061), this means that based on the present evidence, it is possible that some future studies will likely find positive effect.
Finally, Q test has shown a P value above significance level (P=0.2496), which indicates that there is no significant heterogeneity.
Assessment of risk of bias
Overall, there was a low risk of study bias, a table of the included studies with their associated risk of bias is shown in Table S1. One of the main observations was that a significant number of studies (51 out of 81 studies) did not have a comparator conventional test to draw conclusion on the performance of the models compared to current practice. However, the majority of the studies reported reasonable information about their models and performance metrics.
Discussion
Deep learning techniques
The three main types of layers which compose artificial neural networks (ANNs) are: input layers taking the raw image data, hidden layers connected via weight vectors, and an output layer which takes the weighted sum, applies an output function and return a prediction.
The fully connected layers with MLP put significant limitation to the size of the model and the number of filters available to learn image features. CNNs overcome this challenge by using fully connected layers very sparsely, and with more focus on convolution layers using hundreds or thousands of filters, the values of which are learnt automatically during the training phase. The sequential nature of the layers of the CNN can be thought of in the following steps: the early layers detect edges from raw pixel data, these edges are then used to detect shapes in further layers, and these shapes are used to detect higher-level features in the later layers. An additional exciting property of neural networks is that they can be used with transfer learning where high-level feature extraction ability is kept by saving the majority of the network, and a new layer to fit with the purpose of the study is exchanged with the output layer (85).
GANs have been gaining more popularity recently in medical imaging, and we saw some novel applications which have been applied in CCTA and invasive coronary angiography, as shown in Figure 3. These networks were first introduced by Goodfellow et al. (86), and can be used to generate synthetic images that are perceptually similar to their ground truth, authentic originals. This can be achieved by training two neural networks, one is called the generator that accepts an input vector of randomly generated noise and produces an output “imitation” image that looks similar to an image from the training image domain, if not identical to an authentic image, and the other is called the discriminator which attempts to determine if a given image is an “authentic” or “fake”. By training both of these networks at the same time, one giving feedback to the other, we can learn to generate synthetic images. This model has been applied by Du et al. (50) successfully to unravel the complex features of coronary lesions seen in invasive coronary angiography by combining images from lesion location with images from lesion morphology to generate a high-level diagnostic information including identification of every coronary artery lesion and the coronary artery segment, in which it is located.
Finally, RNNs are type of neural networks which uses sequential data or time series data. They are distinguished by their memory as they take information from prior inputs to influence the current output. An RNN cell contains a closed-loop which allows the output of the current step to be influenced by the output of the previous step. Carson et al. (79) applied a RNN on CCTA to predict FFR based on the fact that coronary anatomy geometry has large variations including different vessel sizes, connectivity and the inclusion or exclusion of certain vessels. RNN has the advantage for providing the solution in the next vessel based on the solution of the previous vessel. This model had high performance compared to other non-invasive models and perfect sensitivity when compared with invasive FFR, however, it had very low specificity at 40% with high rate of FP FFR. This study had a small sample size of only 25 cases. Therefore, further testing and studies on RNN is required for further evaluation.
Summary of main results
This systematic review shows how extensive the work has been made in the last few years in the field of coronary anatomy and atherosclerosis imaging using machine learning and deep learning applications. Overall, all studies reported in this review (81 studies over 6 years) showed good performance of the models presented to achieve the target outputs for each individual study.
The most popular imaging modality which has been used extensively in deep learning application is CCTA, with a wide range of applications ranging from coronary anatomy segmentation, plaque classification, coronary calcium quantification, vulnerable plaque detection, noise reduction and image reconstruction, and clinical outcome prediction.
Invasive coronary angiography was a focus in deep learning in recent years, various applications looked at coronary segmentation, coronary stenosis classification, thrombus detection, total occlusion detection and dissection detection. Moreover, the intra-vascular coronary imaging modalities such as IVUS and OCT have been studied for the last few years for various applications, mainly linked to segmentation and characterisation of coronary artery lumen and plaque.
One of the major works, which shows how effective deep learning can be is the CT-FFR algorithm. Our meta-analysis of the 8 studies looking at deep learning applications to predict CT-FFR showed positive results of the pooled diagnostic performance and low level of heterogeneity. Furthermore, predictive interval tests showed that some future studies will likely find positive effect based on the present evidence. Although CT-FFR was performed initially by Itu et al. (6) using a MLP, it gained popularity after showing superior performance to computational fluid dynamics and was tested in several studies for external validation, which confirmed its utility in clinical applications. There is currently more focus on using more advanced deep learning techniques such as CNN, and this continues to show promising results.
The positive findings in all the presented studies could have an impact on clinical practice by introducing new developments to current state of the art imaging modalities, such as CCTA, IVUS, and OCT, and improve clinical workflow with faster diagnosis and more meaningful image analysis.
The quantification ability of deep learning and radiomics can unravel features and relationships in the medical images which are not easily detected by the human eye, however, this area still needs further studies to evaluate the clinical usage of such models, and the current review has set the scene for the potential, which computer vision could offer to achieve this goal.
Limitations
This review excluded studies which have been presented in computer vision competitions, which may underrepresent some of the effective techniques out in the industry, therefore, the list of the models listed here is not exclusive.
The presented studies in this review have reported a large variation of performance metrics, which made meta-analysis challenging and it is limited to only 8 studies.
Conclusions
Implications for practice
This review has shed light on an important rising field in cardiovascular imaging, deep learning and computer vision. The tremendous advancement in coronary atherosclerosis imaging has already affected our practice with the use of non-invasive CT-FFR to make clinical decisions, and will soon change many other decisions we make in cardiovascular medicine. Although this is an exciting era of technology and precision medicine, clinical scrutiny and systematic review of the evidence is essential and should be periodic, in order to make the best possible decision for our patients.
Implications for research
There is a high demand for more research using novel deep learning applications on large datasets, in well-designed environments with robust study protocols, to achieve meaningful software applications, which are trustworthy and reliable to use on our patients.
Acknowledgments
Funding: This research has received grant from Wellcome Trust (222678/Z/21/Z).
Footnote
Reporting Checklist: The authors have completed the PRISMA reporting checklist. Available at https://jmai.amegroups.com/article/view/10.21037/jmai-22-36/rc
Peer Review File: Available at https://jmai.amegroups.com/article/view/10.21037/jmai-22-36/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://jmai.amegroups.com/article/view/10.21037/jmai-22-36/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Nowbar AN, Gitto M, Howard JP, et al. Mortality From Ischemic Heart Disease. Circ Cardiovasc Qual Outcomes 2019;12:e005375. [Crossref] [PubMed]
- Leeflang MM, Deeks JJ, Takwoingi Y, et al. Cochrane diagnostic test accuracy reviews. Syst Rev 2013;2:82. [Crossref] [PubMed]
- Rodrigues ÉO, Morais FF, Morais NA, et al. A novel approach for the automated segmentation and volume quantification of cardiac fats on computed tomography. Comput Methods Programs Biomed 2016;123:109-28. [Crossref] [PubMed]
- Kang D, Dey D, Slomka PJ, et al. Structured learning algorithm for detection of nonobstructive and obstructive coronary plaque lesions from computed tomography angiography. J Med Imaging (Bellingham) 2015;2:014003. [Crossref] [PubMed]
- Araki T, Ikeda N, Shukla D, et al. A new method for IVUS-based coronary artery disease risk stratification: A link between coronary & carotid ultrasound plaque burdens. Comput Methods Programs Biomed 2016;124:161-79. [Crossref] [PubMed]
- Itu L, Rapaka S, Passerini T, et al. A machine-learning approach for computation of fractional flow reserve from coronary computed tomography. J Appl Physiol (1985) 2016;121:42-52. [PubMed]
- Wolterink JM, Leiner T, de Vos BD, et al. Automatic coronary artery calcium scoring in cardiac CT angiography using paired convolutional neural networks. Med Image Anal 2016;34:123-36. [Crossref] [PubMed]
- Su S, Hu Z, Lin Q, et al. An artificial neural network method for lumen and media-adventitia border detection in IVUS. Comput Med Imaging Graph 2017;57:29-39. [Crossref] [PubMed]
- Yong YL, Tan LK, McLaughlin RA, et al. Linear-regression convolutional neural network for fully automated coronary lumen segmentation in intravascular optical coherence tomography. J Biomed Opt 2017;22:1-9. [Crossref] [PubMed]
- Mengdi Xu, Jun Cheng, Annan Li, et al. Fibroatheroma identification in Intravascular Optical Coherence Tomography images using deep features. Annu Int Conf IEEE Eng Med Biol Soc 2017;2017:1501-4. [Crossref] [PubMed]
- Zreik M, van Hamersvelt RW, Wolterink JM, et al. A Recurrent CNN for Automatic Detection and Classification of Coronary Artery Plaque and Stenosis in Coronary CT Angiography. IEEE Trans Med Imaging 2019;38:1588-98. [Crossref] [PubMed]
- Zreik M, Lessmann N, van Hamersvelt RW, et al. Deep learning analysis of the myocardium in coronary CT angiography for identification of patients with functionally significant coronary artery stenosis. Med Image Anal 2018;44:72-85. [Crossref] [PubMed]
- Kolluru C, Prabhu D, Gharaibeh Y, et al. Deep neural networks for A-line-based plaque classification in coronary intravascular optical coherence tomography images. J Med Imaging (Bellingham) 2018;5:044504. [Crossref] [PubMed]
- Zhang L, Wahle A, Chen Z, et al. Predicting Locations of High-Risk Plaques in Coronary Arteries in Patients Receiving Statin Therapy. IEEE Trans Med Imaging 2018;37:151-61. [Crossref] [PubMed]
- Oh SJ, Lee G, Choi T, et al. Detection of vulnerable plaque with deep learning algorithm in IVUS imaging. 2018. Available online: https://abstractbook.pcronline.com/export/pdf/id/100155
- van Rosendael AR, Maliakal G, Kolli KK, et al. Maximization of the usage of coronary CTA derived plaque information using a machine learning based algorithm to improve risk stratification; insights from the CONFIRM registry. J Cardiovasc Comput Tomogr 2018;12:204-9. [Crossref] [PubMed]
- Stuckey TD, Gammon RS, Goswami R, et al. Cardiac Phase Space Tomography: A novel method of assessing coronary artery disease utilizing machine learning. PLoS One 2018;13:e0198603. [Crossref] [PubMed]
- Lessmann N, van Ginneken B, Zreik M, et al. Automatic Calcium Scoring in Low-Dose Chest CT Using Deep Neural Networks With Dilated Convolutions. IEEE Trans Med Imaging 2018;37:615-25. [Crossref] [PubMed]
- Šprem J, de Vos BD, Lessmann N, et al. Impact of automatically detected motion artifacts on coronary calcium scoring in chest computed tomography. J Med Imaging (Bellingham) 2018;5:044007. [Crossref] [PubMed]
- Hae H, Kang SJ, Kim WJ, et al. Machine learning assessment of myocardial ischemia using angiography: Development and retrospective validation. PLoS Med 2018;15:e1002693. [Crossref] [PubMed]
- Dey D, Gaur S, Ovrehus KA, et al. Integrated prediction of lesion-specific ischaemia from quantitative coronary CT angiography using machine learning: a multicentre study. Eur Radiol 2018;28:2655-64. [Crossref] [PubMed]
- van Hamersvelt RW, Zreik M, Voskuil M, et al. Deep learning analysis of left ventricular myocardium in CT angiographic intermediate-degree coronary stenosis improves the diagnostic accuracy for identification of functionally significant stenosis. Eur Radiol 2019;29:2350-9. [Crossref] [PubMed]
- Cho H, Lee JG, Kang SJ, et al. Angiography-Based Machine Learning for Predicting Fractional Flow Reserve in Intermediate Coronary Artery Lesions. J Am Heart Assoc 2019;8:e011685. [Crossref] [PubMed]
- Liu R, Zhang Y, Zheng Y, et al. Automated Detection of Vulnerable Plaque for Intravascular Optical Coherence Tomography Images. Cardiovasc Eng Technol 2019;10:590-603. [Crossref] [PubMed]
- Gessert N, Lutz M, Heyder M, et al. Automatic Plaque Detection in IVOCT Pullbacks Using Convolutional Neural Networks. IEEE Trans Med Imaging 2019;38:426-34. [Crossref] [PubMed]
- Abdolmanafi A, Dahdah N, Duong L, et al. Fully automatic artificial intelligence diagnostic model of coronary artery lesions using oct imaging. Can J Cardiol 2019;35:S61-2. [Crossref]
- Liu X, Yang R, Xie L, et al. TCT-242 Detection and Classification of Coronary Bifurcation Lesions by Using Artificial Intelligence. J Am Coll Cardiol 2019;74:B241. [Crossref]
- Gharaibeh Y, Prabhu D, Kolluru C, et al. Coronary calcification segmentation in intravascular OCT images using deep learning: application to calcification scoring. J Med Imaging (Bellingham) 2019;6:045002. [Crossref] [PubMed]
- Jun TJ, Kang SJ, Lee JG, et al. Automated detection of vulnerable plaque in intravascular ultrasound images. Med Biol Eng Comput 2019;57:863-76. [Crossref] [PubMed]
- Lee PC, Lee N, Pyo R. Convolutional Neural Networks for Interpretation of Coronary Angiography. Circulation 2019;140:A12950.
- Yang S, Kweon J, Roh JH, et al. Deep learning segmentation of major vessels in X-ray coronary angiography. Sci Rep 2019;9:16897. [Crossref] [PubMed]
- Wang YY, Qiu CH, Jiang J, et al. Detecting the Media-adventitia Border in Intravascular Ultrasound Images through a Classification-based Approach. Ultrason Imaging 2019;41:78-93. [Crossref] [PubMed]
- Johnson KM, Johnson HE, Zhao Y, et al. Scoring of Coronary Artery Disease Characteristics on Coronary CT Angiograms by Using Machine Learning. Radiology 2019;292:354-62. [Crossref] [PubMed]
- Kolossváry M, Karády J, Kikuchi Y, et al. Radiomics versus Visual and Histogram-based Assessment to Identify Atheromatous Lesions at Coronary CT Angiography: An ex Vivo Study. Radiology 2019;293:89-96. [Crossref] [PubMed]
- Wang ZQ, Zhou YJ, Zhao YX, et al. Diagnostic accuracy of a deep learning approach to calculate FFR from coronary CT angiography. J Geriatr Cardiol 2019;16:42-8. [PubMed]
- Datong C, Minghui L, Cheng J, et al. Coronary Calcium Detection Based on Improved Deep Residual Network in Mimics. J Med Syst 2019;43:119. [Crossref] [PubMed]
- Oikonomou EK, Williams MC, Kotanidis CP, et al. A novel machine learning-derived radiotranscriptomic signature of perivascular fat improves cardiac risk prediction using coronary CT angiography. Eur Heart J 2019;40:3529-43. [Crossref] [PubMed]
- Masuda T, Nakaura T, Funama Y, et al. Machine-learning integration of CT histogram analysis to evaluate the composition of atherosclerotic plaques: Validation with IB-IVUS. J Cardiovasc Comput Tomogr 2019;13:163-9. [Crossref] [PubMed]
- Kigka VI, Sakellarios AI, Tsompou P, et al. Site specific prediction of atherosclerotic plaque progression using computational biomechanics and machine learning. Annu Int Conf IEEE Eng Med Biol Soc 2019;2019:6998-7001. [Crossref] [PubMed]
- Zhang L, Mayrhofer T, Foldyna B, et al. Machine Learning for Prediction of Cardiovascular Outcomes in Symptomatic Outpatients With Suspected Coronary Artery Disease Having Coronary CT Angiography: A Secondary Analysis of the Promise Trial. Circulation 2019;140:A14038.
- Commandeur F, Goeller M, Razipour A, et al. Fully Automated CT Quantification of Epicardial Adipose Tissue by Deep Learning: A Multicenter Study. Radiol Artif Intell 2019;1:e190045. [Crossref] [PubMed]
- Hong Y, Commandeur F, Cadet S, et al. Deep learning-based stenosis quantification from coronary CT Angiography. Proc SPIE Int Soc Opt Eng 2019;10949:109492I.
- Huo Y, Terry JG, Wang J, et al. Coronary Calcium Detection using 3D Attention Identical Dual Deep Network Based on Weakly Supervised Learning. Proc SPIE Int Soc Opt Eng 2019;10949:1094917.
- Wang L, Tang D, Maehara A, et al. Using intravascular ultrasound image-based fluid-structure interaction models and machine learning methods to predict human coronary plaque vulnerability change. Comput Methods Biomech Biomed Engin 2020;23:1267-76. [Crossref] [PubMed]
- Lee JG, Ko J, Hae H, et al. Intravascular ultrasound-based machine learning for predicting fractional flow reserve in intermediate coronary artery lesions. Atherosclerosis 2020;292:171-7. [Crossref] [PubMed]
- Wu W, Zhang J, Xie H, et al. Automatic detection of coronary artery stenosis by convolutional neural network with temporal constraint. Comput Biol Med 2020;118:103657. [Crossref] [PubMed]
- Sampedro-Gómez J, Dorado-Díaz PI, Vicente-Palacios V, et al. Machine Learning to Predict Stent Restenosis Based on Daily Demographic, Clinical, and Angiographic Characteristics. Can J Cardiol 2020;36:1624-32. [Crossref] [PubMed]
- Miyoshi T, Higaki A, Kawakami H, et al. Automated interpretation of the coronary angioscopy with deep convolutional neural networks. Open Heart 2020;7:e001177. [Crossref] [PubMed]
- Zhang D, Yang G, Zhao S, et al. Direct Quantification of Coronary Artery Stenosis Through Hierarchical Attentive Multi-View Learning. IEEE Trans Med Imaging 2020;39:4322-34. [Crossref] [PubMed]
- Du T, Xie L, Zhang H, et al. Training and validation of a deep learning architecture for the automatic analysis of coronary angiography. EuroIntervention 2021;17:32-40. [Crossref] [PubMed]
- He C, Wang J, Yin Y, et al. Automated classification of coronary plaque calcification in OCT pullbacks with 3D deep neural networks. J Biomed Opt 2020;25:095003. [Crossref] [PubMed]
- Yabushita H, Goto S, Nakamura S, et al. Development of Novel Artificial Intelligence to Detect the Presence of Clinically Meaningful Coronary Atherosclerotic Stenosis in Major Branch from Coronary Angiography Video. J Atheroscler Thromb 2021;28:835-43. [Crossref] [PubMed]
- Hamaya R, Hoshino M, Yonetsu T, et al. Defining heterogeneity of epicardial functional stenosis with low coronary flow reserve by unsupervised machine learning. Heart Vessels 2020;35:1527-36. [Crossref] [PubMed]
- Lee J, Prabhu D, Kolluru C, et al. Automated plaque characterization using deep learning on coronary intravascular optical coherence tomographic images. Biomed Opt Express 2019;10:6497-515. [Crossref] [PubMed]
- Min HS, Yoo JH, Kang SJ, et al. Detection of optical coherence tomography-defined thin-cap fibroatheroma in the coronary artery using deep learning. EuroIntervention 2020;16:404-12. [Crossref] [PubMed]
- Commandeur F, Slomka PJ, Goeller M, et al. Machine learning to predict the long-term risk of myocardial infarction and cardiac death based on clinical risk, coronary calcium, and epicardial adipose tissue: a prospective study. Cardiovasc Res 2020;116:2216-25. [Crossref] [PubMed]
- Muscogiuri G, Chiesa M, Trotta M, et al. Performance of a deep learning algorithm for the evaluation of CAD-RADS classification with CCTA. Atherosclerosis 2020;294:25-32. [Crossref] [PubMed]
- Benz DC, Benetos G, Rampidis G, et al. Validation of deep-learning image reconstruction for coronary computed tomography angiography: Impact on noise, image quality and diagnostic accuracy. J Cardiovasc Comput Tomogr 2020;14:444-51. [Crossref] [PubMed]
- Wang W, Wang H, Chen Q, et al. Coronary artery calcium score quantification using a deep-learning algorithm. Clin Radiol 2020;75:237.e11-6. [Crossref] [PubMed]
- Al'Aref SJ, Maliakal G, Singh G, et al. Machine learning of clinical variables and coronary artery calcium scoring for the prediction of obstructive coronary artery disease on coronary computed tomography angiography: analysis from the CONFIRM registry. Eur Heart J 2020;41:359-67. [Crossref] [PubMed]
- Kawasaki T, Kidoh M, Kido T, et al. Evaluation of Significant Coronary Artery Disease Based on CT Fractional Flow Reserve and Plaque Characteristics Using Random Forest Analysis in Machine Learning. Acad Radiol 2020;27:1700-8. [Crossref] [PubMed]
- Fischer AM, Eid M, De Cecco CN, et al. Accuracy of an Artificial Intelligence Deep Learning Algorithm Implementing a Recurrent Neural Network With Long Short-term Memory for the Automated Detection of Calcified Plaques From Coronary Computed Tomography Angiography. J Thorac Imaging 2020;35:S49-57. [Crossref] [PubMed]
- van Velzen SGM, Lessmann N, Velthuis BK, et al. Deep Learning for Automatic Calcium Scoring in CT: Validation Using Multiple Cardiac CT and Chest CT Protocols. Radiology 2020;295:66-79. [Crossref] [PubMed]
- Zreik M, van Hamersvelt RW, Khalili N, et al. Deep Learning Analysis of Coronary Arteries in Cardiac CT Angiography for Detection of Patients Requiring Invasive Coronary Angiography. IEEE Trans Med Imaging 2020;39:1545-57. [Crossref] [PubMed]
- Kumamaru KK, Fujimoto S, Otsuka Y, et al. Diagnostic accuracy of 3D deep-learning-based fully automated estimation of patient-level minimum fractional flow reserve from coronary computed tomography angiography. Eur Heart J Cardiovasc Imaging 2020;21:437-45. [PubMed]
- Candemir S, White RD, Demirer M, et al. Automated coronary artery atherosclerosis detection and weakly supervised localization on coronary CT angiography with a deep 3-dimensional convolutional neural network. Comput Med Imaging Graph 2020;83:101721. [Crossref] [PubMed]
- Shu ZY, Cui SJ, Zhang YQ, et al. Predicting Chronic Myocardial Ischemia Using CCTA-Based Radiomics Machine Learning Nomogram. J Nucl Cardiol 2022;29:262-74. [Crossref] [PubMed]
- van den Oever LB, Cornelissen L, Vonder M, et al. Deep learning for automated exclusion of cardiac CT examinations negative for coronary artery calcium. Eur J Radiol 2020;129:109114. [Crossref] [PubMed]
- Han D, Liu J, Sun Z, et al. Deep learning analysis in coronary computed tomographic angiography imaging for the assessment of patients with coronary artery stenosis. Comput Methods Programs Biomed 2020;196:105651. [Crossref] [PubMed]
- Han D, Kolli KK, Al'Aref SJ, et al. Machine Learning Framework to Identify Individuals at Risk of Rapid Progression of Coronary Atherosclerosis: From the PARADIGM Registry. J Am Heart Assoc 2020;9:e013958. [Crossref] [PubMed]
- Lin A, Kolossváry M, Yuvaraj J, et al. Myocardial Infarction Associates With a Distinct Pericoronary Adipose Tissue Radiomic Phenotype: A Prospective Case-Control Study. JACC Cardiovasc Imaging 2020;13:2371-83. [Crossref] [PubMed]
- Chen M, Wang X, Hao G, et al. Diagnostic performance of deep learning-based vascular extraction and stenosis detection technique for coronary artery disease. Br J Radiol 2020;93:20191028. [Crossref] [PubMed]
- Tesche C, Bauer MJ, Baquet M, et al. Improved long-term prognostic value of coronary CT angiography-derived plaque measures and clinical parameters on adverse cardiac outcome using machine learning. Eur Radiol 2021;31:486-93. [Crossref] [PubMed]
- Al'Aref SJ, Singh G, Choi JW, et al. A Boosted Ensemble Algorithm for Determination of Plaque Stability in High-Risk Patients on Coronary CTA. JACC Cardiovasc Imaging 2020;13:2162-73. [Crossref] [PubMed]
- Hong JH, Park EA, Lee W, et al. Incremental Image Noise Reduction in Coronary CT Angiography Using a Deep Learning-Based Technique with Iterative Reconstruction. Korean J Radiol 2020;21:1165-77. [Crossref] [PubMed]
- Podgorsak AR, Sommer KN, Reddy A, et al. Initial evaluation of a convolutional neural network used for noninvasive assessment of coronary artery disease severity from coronary computed tomography angiography data. Med Phys 2020;47:3996-4004. [Crossref] [PubMed]
- Eberhard M, Nadarevic T, Cousin A, et al. Machine learning-based CT fractional flow reserve assessment in acute chest pain: first experience. Cardiovasc Diagn Ther 2020;10:820-30. [Crossref] [PubMed]
- Son J, Shin JY, Chun EJ, et al. Predicting High Coronary Artery Calcium Score From Retinal Fundus Images With Deep Learning Algorithms. Transl Vis Sci Technol 2020;9:28. [Crossref] [PubMed]
- Carson JM, Chakshu NK, Sazonov I, et al. Artificial intelligence approaches to predict coronary stenosis severity using non-invasive fractional flow reserve. Proc Inst Mech Eng H 2020;234:1337-50. [Crossref] [PubMed]
- Gangl C, Roth C, Dalos D, et al. P5627 Automated detection of calcified plaques in coronary optical coherence tomography images using image segmentation based on machine learning. Eur Heart J 2019;40:ehz746.0571.
- Głowacki J, Krysiński M, Czaja-Ziółkowska M, et al. Machine Learning-based Algorithm Enables the Exclusion of Obstructive Coronary Artery Disease in the Patients Who Underwent Coronary Artery Calcium Scoring. Acad Radiol 2020;27:1416-21. [Crossref] [PubMed]
- Hoshino M, Sugiyama T, Kanaji Y, et al. Prognostic value of peri-coronary adipose tissue attenuation and phenotypic clustering of whole vessel and lesion plaque quantification on coronary computed tomography angiography. J Am Coll Cardiol 2020;75:1805. [Crossref]
- Kawaguchi YO, Fujimoto S, Kumamaru KK, et al. Fully automated 3D deep-learning analysis of coronary CT Angiography: Prediction of fractional flow reserve. Circulation 2018;138:A12206.
- Blackman NJ. Systematic reviews of evaluations of diagnostic and screening tests. Odds ratio is not independent of prevalence. BMJ 2001;323:1188. [Crossref] [PubMed]
- Rosebrock A. Deep learning for computer vision. 2019. Available online: https://pyimagesearch.com/deep-learning-computer-vision-python-book/
- Goodfellow IJ, Pouget-Abadie J, Mirza M, et al. Generative adversarial networks. arXiv preprint. arXiv:1406.2661.
Cite this article as: Alskaf E, Dutta U, Scannell CM, Chiribiri A. Deep learning applications in coronary anatomy imaging: a systematic review and meta-analysis. J Med Artif Intell 2022;5:11.