Skin cancer detection using multi-scale deep learning and transfer learning
Highlight box
Key findings
• A new model was designed to detect skin cancer lesions.
What is known and what is new?
• Deep learning is among the best methods for skin cancer detection.
• Using different techniques for data augmentation.
What is the implication, and what should change now?
• Sometimes a simple deep learning structure is not enough in complicated problems. Based on problem new structures and techniques should be used.
Introduction
Background
The largest body organ is the skin, so it is not strange that skin cancer has the highest rate among human compared to other cancer types (1,2). The skin cancers are divided into two parts, melanoma and non-melanoma (3). Similar to other cancers, it is important to diagnose the cancer in the early stages. Melanoma is the deadliest type of skin cancer (4). It happens when melanocytes cells grow and become out of control. It can happen in any part of the body, but most in parts that are exposed to sunlight (5). The only way that the skin cancer can be detected now is to use biopsy, which is removing sample from the skin part to determine if it is cancerous or not (5). Due to medical mistakes (6) that sometimes happens, it is wise to use automatic detection method to help physicians to detect the cancer as soon as possible. Using machine learning methods, the process of detecting the cancer can be much faster. In recent decade, deep learning is considered to be one of the best methods in detection and recognition (5).
Rationale and knowledge gap
Data is the most important part in machine learning methods. More data can help to train the learning model better and reduce overfitting. When the model cannot generalize well, and it fits completely or closely to the training data, overfitting happens. Usually, there are not enough data in learning problems due to difficulties in creating the datasets, especially when the data are images. The data needs to be labeled, which takes a long time for large datasets. For this reason, one of the main tasks in each learning problem is how to increase the number of images in the dataset. Many methods to increase the images have been discussed in the paper.
Feature extraction is the second important part of a classification system. By feature extraction, one tries to extract the most important information from the data. In deep learning structures, the feature extraction and classification part are done in one phase. When working on images, it is important to extract the features from the areas that there is higher probability to contain the information that we are looking for. In skin cancer datasets, most of the skin parts that needs attention are located at the center of the image (refer to Figure 1). This fact has been used in this paper in order to improve the feature extraction part. Three different convolutional neural networks (CNNs) are trained and the output of each are combined and feed to a fully connected network (FCN). Results show improvement in the detection rate based on this combination.
This paper is subdivided into four different parts. The “Methods” section describes the literature review, the methodology that was used and the idea of using deep learning and transfer learning is described in detail. In the “Results” section, the results and comparison with other methods are presented. The “Discussion” section states the conclusion and presents some ideas for future works. The author presents this article in accordance with the TRIPOD reporting checklist (available at https://jmai.amegroups.com/article/view/10.21037/jmai-23-67/rc).
Methods
Literature review
Different researchers have worked on skin cancer problem. In this part, we will have a quick review of different methods that have used so far. Ali et al. used LightNet for classification. Their model was suited for mobile applications. The tests were applied over International Skin Imaging Collaboration (ISIC) 2016 datasets and the results were published based on accuracy (81.60%), sensitivity (14.90%), and specificity (98.00%) (8). Dorj et al. used support vector machine (SVM) with deep SVM to diagnose different types of skin cancers such as basal cell carcinoma (BCC), squamous cell carcinoma (SCC), and melanoma. BCC and SCC are among non-melanoma cancers that do not spread to other parts and be detected much easier compare to melanoma cancers (9). Their algorithm was tested on 3,753 images, which include four kinds of skin cancers images. The best results that they got for accuracy, sensitivity, and specificity were 95.1% (SCC), 98.9% (actinic keratosis), and 94.17% (SCC), respectively. Minimum values were 91.8% (BCC), 96.9% (SCC), and 90.74% (melanoma), respectively (9).
Esteva used CNN to classify the images into three categories Melanoma, benign Keratinocyte carcinomas and benign seborrheic keratosis (SK) (10). In reference (11), a hybrid system based on SVM, sparse coding and deep learning method were used for skin cancer detection. They used ISIC, which contained 2,624 clinical cases of melanoma [334], atypical nevi [144], and benign lesions [2,146]. Two-fold cross-validation was used 20 times for evaluation, and for classification two tasks were presented: (I) melanoma vs. all non-melanoma lesions, and (II) melanoma vs. atypical lesions only. For the first task an accuracy of 93.1%, 94.9% recall, and 92.8% specificity, and for the second task 73.9% accuracy, 73.8% recall, and 74.3% specificity.
Harangi et al. used CNN combining of AlexNet, VGGNet, and GoogleNetfor structures (12). Kalouche used CNN and focused on edges to detect skin cancer (13). Rezvantalab et al. showed that deep learning models outperforms other models by at least 11%, DenseNet201, Inception v3, ResNet 152, and InceptionResNet v2 were used (14). Sagar et al. used ResNet-50 with deep transfer learning over 3,600 lesion images that were chosen from ISIC dataset. The proposed model showed better performance than InceptionV3, DenseNet169 (15). In Jojoa Acosta et al. work region of interest was extracted by mask and region-based CNN, and ResNet152 was used for classification. 2742 images from ISIC were used (16).
Some other papers were also published based on generative adversarial network (GAN) and progressive GAN (PGAN). Rashid et al. used GAN on ISIC 2018 dataset. They tried to diagnose benign keratosis, dermatofibroma, melanoma, melanocytic nevus, vascular lesion, etc. Deconvolutional network was used, and they reached to 86.1% accuracy (17). Bisla et al. used deep convolutional GAN and decoupled deep convolutional GAN for data augmentation. ISIC 2017 and ISIC 2018 were among the datasets that were used. They reached to 86.1% accuracy (18). Ali et al. used self-attention based PGAN to detect vascular, pigmented benign keratosis, pigmented Bowen’s, nevus, dermatofibroma, etc. ISIC 2018 dataset was used. A generative model was enhanced with a stabilization technique. The accuracy was 70.1% (19). Zunair et al. designed a two-stage framework for automatic classification of skin lesion images. Adversarial training and transfer learning were used reference (20). A list of other related papers can be found in reference (5).
Methodology
In the next two subsections, the proposed method is discussed. In the first part, the augmentation and image processing part are described. The model is described in the second subsection.
Preprocessing
Preprocessing is an important part of medical images. Two different tasks should be considered in preprocessing. One is an image preparation part that fix some color issues or enhance the image and the other one is data augmentation part in which the number of images in the dataset is increased in different ways. The first step was to normalize the images. This was done by subtracting the mean of the red, green, blue (RGB) value from the images in the dataset. This method was suggested by Krizhevsky et al. (21). For building a strong CNN many images are needed for training and testing which is a huge challenge for skin cancer detection problem. Creating large datasets with variety in different cancer types is time-consuming. The data set that is used for this research is ISIC 2020 (7,22). It contains, 33,126 dermoscopic training images from 2,000 patients. In total, 1,400 (around 4%) of these images were chosen for testing and set aside. Only the training set is augmented to provide more examples to the model and increase its robustness (while the test set does not need to be augmented, since real images are needed to test performance). In order to increase the number of training images, each image was rotated 10 degrees which resulted in 35×31,726 new images.
Transfer learning
Training CNN is not an easy task, the reason is CNN needs many images for training and testing. Transfer learning is used to overcome the problem of not having many images in addition to image enhancement. In transfer learning, a network will use the weights of a neural network that was trained on another image dataset similar to what we want to exploit, and the dataset have had enough samples. In this work, transfer learning was exploited as follows: First, the CNN weights for ImageNet (23) are used as the initial weights for the network. It means that a CNN was fully trained by using the ImageNet dataset. In this network, the last layer (the last layer of the fully connected layer) that is designed for classification of 1,000 classes (number of classes in ImageNet) is changed with a softmax layer with two classes (melanoma vs. non-melanoma), which is binary classification (just to mention instead of softmax sigmoid function can also be used). In softmax layer, the number of nodes is equal to the number of outputs, and the value shows the probability of each output. In the next step, back-propagation (a method that tries to adjust the weights based on the error in the output) is applied to the network to fine-tune the weights, so the network become more compatible with the new classes. What is important is not allowing the weights to be changed dramatically, so a small learning rate should be used. Learning rate, is a hyperparameter used in neural network to control the change in model based on the error. By using a high learning rate, the weights will be totally ruined. Applying the back-propagation to a certain point can be considered as a hyperparameter in the training process.
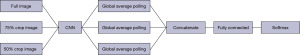
Another technique that is used in this paper, is similar to the idea used in (24) but the structure used is more advanced and is combined with transfer learning. Three different networks were trained based on different images and transfer learning. One, for the images with the real size, which means to apply the whole process described above over the skin cancer dataset. In skin cancer detection problem, the area that is cancerous is in the middle of the image and most of the surrounding part belongs to the skin part that is not cancerous and may mislead the network. There are two ways to overcome this problem. One is segmentation (25,26) and the other is the proposed method. Segmentation divides the image into two parts, the cancerous part and other part of the image, but it has its own drawbacks. The main drawback is that the segmentation problem has errors which will affect the learning. In the proposed method, two other networks were trained. One for the images with cropping 50% of the original image and then resizing it back to the original size and the third one for the images with cropping 75% of the original image and resize it. Figures 1-3 show the images with original size, 75%, and 50% crop.
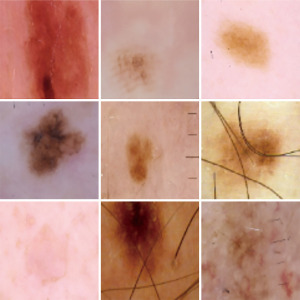
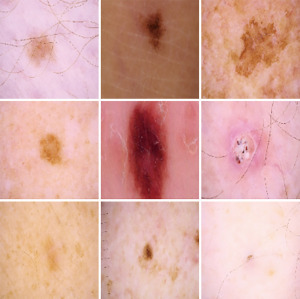
The three networks were trained and fine-tuned. Then the information from the global-average-pooling-two-dimensional (2D) (pooling layer reduces the dimensions and finds the highest response in local parts of the feature map) were combined to create a vector based on the information extracted from the three structures. A fully connected layer was created with two outputs. Figure 4 shows the whole structure.
The value for number of batches, learning rate, and epochs were 32, 10, and 0.0001, and the learning rate for fine-tuning was 0.00001. The CNN used was MobileNetV2, a CNN with 53 layers.
The performance of the proposed structure is measured with the following measures.
Where TP, TN, FP, and FN stand for true positive, true negative, false positive, and false negative, respectively.
Results
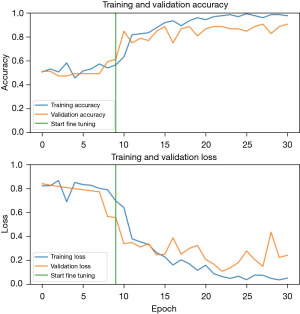
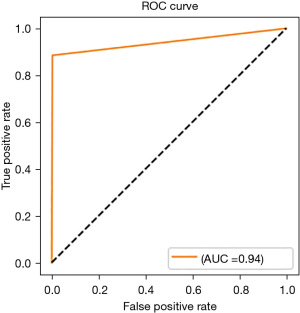
The experiments were done on Lambda laptop, with GPU NVIDA RTX 3080 Max-Q, VRAM 16 GB GDDR6, CPU Intel Core i7-1 1800H, and RAM 64 GB 3200 MHZ DDR4. Figure 5 shows the accuracy and loss of the proposed method and Figure 6 shows the receiver operating characteristic (ROC) curve.
Figure 5 shows the plot for accuracy and loss. Table 1 shows the result for the proposed method comparing to some other methods, and Table 2 shows the confusion matrix. It is worth mentioning that the test set for some methods may differ from each other.
Table 1
| Method | Recall (%) | Precision (%) | Accuracy (%) | Dataset |
|---|---|---|---|---|
| (8) | 14.9 | – | 81.6 | ISIC 2016 |
| (24) | – | – | 90.3 | ISIC |
| (27) | 81 | 75 | 81 | 170 skin lesion images |
| (15) | 77 | 94 | 93.5 | 3,600 lesion images from the ISIC dataset |
| (16) | 82 | – | 90.4 | 2,742 dermoscopic images from ISIC dataset |
| Proposed method | 88.5 | 91.7 | 94.42 | ISIC 2020 |
ISIC, International Skin Imaging Collaboration.
Table 2
| Predicted value/actual value | Cancer | Non-cancer |
|---|---|---|
| Cancer | 354 | 46 |
| Non-cancer | 32 | 968 |
Discussion
The results show that the proposed method outperform other methods in accuracy and recall, also it is comparable to other methods in case of precision. Based on the proposed structure the model has more focus on the cancerous part of the image compare to using a simple deep learning model. Another achievement in this work was using different techniques of image augmentation to increase the number of images in the training set. Applying this technique with the aid of transfer learning increased the detection rate compared to other methods.
The only drawback that our method has is it takes more time if the program is used in sequential way. Using parallel programming will omit the drawback and it will be as fast as other methods.
Conclusions
In this paper, a multi-scale structure was designed and implemented for skin cancer detection. The system was capable of detecting the melanoma vs. non-melanoma. The accuracy was improved by using some image processing techniques to increase the quality and quantity of the training images. By using a multi-scale structure, the performance was increased compared to other methods. For the future other experiments can be done using other deep learning structures. Some other image processing techniques can also be applied such as removing the hair and marks from the images. There are different methods, among them deep learning methods has shown better results (28).
Acknowledgments
The data that support the findings of this study are available from the corresponding website (https://challenge2020.isic-archive.com/).
Funding: None.
Footnote
Reporting Checklist: The author has completed the TRIPOD reporting checklist. Available at https://jmai.amegroups.com/article/view/10.21037/jmai-23-67/rc
Data Sharing Statement: Available at https://jmai.amegroups.com/article/view/10.21037/jmai-23-67/dss
Peer Review File: Available at https://jmai.amegroups.com/article/view/10.21037/jmai-23-67/prf
Conflicts of Interest: The author has completed the ICMJE uniform disclosure form (available at https://jmai.amegroups.com/article/view/10.21037/jmai-23-67/coif). The author has no conflicts of interest to declare.
Ethical Statement: The author is accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Ashraf R, Afzal S, Rehman AU, et al. Region-of-interest based transfer learning assisted framework for skin cancer detection. IEEE Access 2020;8:147858-71.
- Byrd AL, Belkaid Y, Segre JA. The human skin microbiome. Nat Rev Microbiol 2018;16:143-55. [Crossref] [PubMed]
- Elgamal M. Automatic skin cancer images classification. International Journal of Advanced Computer Science and Applications 2013;4:287-94.
- American Cancer Society. Key Statistics for Melanoma Skin Cancer. 2021. Available online: https://www.cancer.org/content/dam/CRC/PDF/Public/8823.00.pdf
- Dildar M, Akram S, Irfan M, et al. Skin Cancer Detection: A Review Using Deep Learning Techniques. Int J Environ Res Public Health 2021;18:5479. [Crossref] [PubMed]
-
Skin Cancer Misdiagnosis - International Skin Imaging Collaboration. SIIM-ISIC 2020 Challenge Dataset. 2020. Available online: https://doi.org/
10.34970/2020-ds01 - Ali AA, Al-Marzouqi H. Melanoma detection using regular convolutional neural networks. In: 2017 International Conference on Electrical and Computing Technologies and Applications (ICECTA). IEEE; 2017:1-5.
- Dorj UO, Lee KK, Choi JY, et al. The skin cancer classification using deep convolutional neural network. Multimed Tools Appl 2018;77:9909-24.
- Esteva A, Kuprel B, Novoa RA, et al. Dermatologist-level classification of skin cancer with deep neural networks. Nature 2017;542:115-8. Erratum in: Nature 2017;546:686. [Crossref] [PubMed]
- Codella N, Cai J, Abedini M, et al. Deep learning, sparse coding, and SVM for melanoma recognition in dermoscopy images. In: International workshop on machine learning in medical imaging. Cham: Springer International Publishing; 2015:118-26.
- Harangi B, Baran A, Hajdu A. Classification Of Skin Lesions Using An Ensemble Of Deep Neural Networks. Annu Int Conf IEEE Eng Med Biol Soc 2018;2018:2575-8. [Crossref] [PubMed]
- Kalouche S. Vision-Based Classification of Skin Cancer Using Deep Learning. 2016. (Accessed on 10 January 2021). Available online: https://www.semanticscholar.org/paper/Vision-Based-Classification-of-Skin-Cancer-using Kalouche/b57ba909756462d812dc20fca157b3972bc1f533
- Rezvantalab A, Safigholi H, Karimijeshni S. Dermatologist level dermoscopy skin cancer classification using different deep learning convolutional neural networks algorithms. arXiv:1810.10348. 2018. Available online: https://arxiv.org/abs/1810.10348
- Sagar A, Dheeba J. Convolutional neural networks for classifying melanoma images. BioRxiv: 2020.05. 22.110973. 2020. Available online: https://www.biorxiv.org/content/10.1101/2020.05.22.110973v3.abstract
- Jojoa Acosta MF, Caballero Tovar LY, Garcia-Zapirain MB, et al. Melanoma diagnosis using deep learning techniques on dermatoscopic images. BMC Med Imaging 2021;21:6. [Crossref] [PubMed]
- Rashid H, Tanveer MA, Khan HA. Skin lesion classification using GAN based data augmentation. In: 2019 41st Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC). IEEE; 2019:916-9.
- Bisla D, Choromanska A, Stein JA, et al. Towards automated melanoma detection with deep learning: data purification and augmentation. arXiv:1902.06061. 2019. (Accessed on 10 February 2021). Available online: http://arxiv.org/abs/1902.06061
- Ali IS, Mohamed MF, Mahdy YB. Data Augmentation for Skin Lesion using Self-Attention based Progressive Generative Adversarial Network. arXiv:1910.11960. 2019. (Accessed on 22 January 2021). Available online: http://arxiv.org/abs/1910.11960
- Zunair H, Ben Hamza A. Melanoma detection using adversarial training and deep transfer learning. Phys Med Biol 2020;65:135005. [Crossref] [PubMed]
- Krizhevsky A, Sutskever I, Hinton GE. Imagenet classification with deep convolutional neural networks. In: Advances in Neural Information Processing Systems. 2012:1097-105.
- Rotemberg V, Kurtansky N, Betz-Stablein B, et al. A patient-centric dataset of images and metadata for identifying melanomas using clinical context. Sci Data 2021;8:34. [Crossref] [PubMed]
- Deng J, Dong W, Socher R, et al. Imagenet: A large-scale hierarchical image database. In: 2009 IEEE Conference on Computer Vision and Pattern Recognition. IEEE; 2009:248-55.
- DDeVries T, Ramachandram D. Skin lesion classification using deep multi-scale convolutional neural networks. arXiv:1703.01402. 2017. Available online: https://arxiv.org/abs/1703.01402
- Verma R, Kumar N, Patil A, et al. MoNuSAC2020: A multi-organ nuclei segmentation and classification challenge. IEEE Trans Med Imaging 2021;40:3413-23. [Crossref] [PubMed]
- Hamza AB, Krim H. A variational approach to maximum a posteriori estimation for image denoising. In: International Workshop on Energy Minimization Methods in Computer Vision and Pattern Recognition. Berlin, Heidelberg: Springer Berlin Heidelberg; 2001:19-34.
- Nasr-Esfahani E, Samavi S, Karimi N, et al. Melanoma detection by analysis of clinical images using convolutional neural network. In: 2016 38th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC). IEEE; 2016:1373-6.
- Talavera-Martinez L, Bibiloni P, Gonzalez-Hidalgo M. Hair segmentation and removal in dermoscopic images using deep learning. IEEE Access 2020;9:2694-704.
Cite this article as: Hajiarbabi M. Skin cancer detection using multi-scale deep learning and transfer learning. J Med Artif Intell 2023;6:23.