
Artificial intelligence and clinical stability after the Norwood operation
Highlight box
Key findings
• Combined artificial intelligence (AI) model including convolutional neural network (CNN) can accurately detect changes from critically ill to stable following the Norwood procedure.
What is known and what is new?
• Clinicians rely on a variety of data sources when managing the post-Norwood patient in the intensive care unit (ICU). There is very limited ability to detect or quantify subtle changes (both positive and negative) in a patient’s clinical status over time;
• Clinical Stability Score (CSS) derived from application of CNN and other traditional machine learning (ML) models to critical care data and waveforms may enhance post operative patient monitoring and management.
What is the implication, and what should change now?
• Data, and knowledge exists in the critical care waveforms, and we should utilize it to deploy real time measure of clinical wellness.
Introduction
Postoperative management of the neonate following the Norwood operation is among the most complex and challenging clinical in pediatric critical care. Despite advancements, one-year mortality following the Norwood operation remains 26–36% (1). Following the Norwood operation with systemic-to-pulmonary artery or ventricle-to-pulmonary artery shunt, the patient has an intensive care unit (ICU) and hospital course that is extremely variable in terms of length and cost (2). Therefore, an opportunity exists to develop clinical decision support that improves mortality and decreases utilization of resources, both in terms of cost and inpatient capacity.
Clinicians rely on a variety of data sources when managing the post-Norwood patient in the ICU (3). Continuous monitoring of electrocardiogram (ECG), invasive arterial blood pressure (ABP), central venous pressure or right atrial pressure (CVP), pulse oximetry [peripheral blood oxygen saturation (SpO2)], cerebral near-infrared spectroscopy (NIRS), and multiple temperature sites is common. Intermittent data such as laboratory values, medications administered, ventilator settings, advanced diagnostic tests, and clinical observations are also important. Given the velocity and volume of data generated for each patient, there is risk of failing to detect subtle changes in data that may be revealing clinical progress or setbacks (4,5). Diagnostic errors and safety events are common in modern ICUs and may be preventable with improvements in the environment of care or cognitive aids for providers (6-9).
Artificial intelligence (AI) and machine learning (ML) for healthcare applications are exploding areas of medical research that hold promise in improving patient outcomes, decreasing costs, and improving utilization of scarce healthcare resources (10-12). Convolutional neural networks (CNNs) are a type of deep learning model that are particularly well-suited to recognize patterns in images and waveforms (13). For high-risk patient populations, a great deal of effort is focused solely on the prevention of significant clinical deterioration events. There is very limited ability to detect or quantify subtle changes in clinical status (both positive and negative) in a patient’s clinical status over time. The primary outcome of this study was to derive a model utilizing a CNN algorithm applied to continuous waveforms to accurately detect changes in a patient’s clinical status over the period of their ICU stay and compare this algorithm to traditional ML techniques such as logistic regression (LR) and random forests (RFs) that are mostly applicable to discrete clinical data. The secondary outcome of this study was to retrospectively compare the CNN model to a clinical evaluation of the patient status. We present this article in accordance with the STARD reporting checklist (available at https://jmai.amegroups.com/article/view/10.21037/jmai-22-35/rc).
Methods
The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by Children’s Healthcare of Atlanta (CHOA IRB #372) and Georgia Institute of Technology (GA Tech IRB #H18163) Institutional Review Boards and a waiver of informed consent was granted as all data would be retrospective and de-identified. All patients who underwent the Norwood operation at Children’s Healthcare of Atlanta between October 1, 2016 and September 30, 2019, were included for analysis during their first post-operative ICU course (n=55, Table 1). Of the 55 patients, 40 completed a usual ICU course with complete data, 3 had >1 day of missing waveform data, 6 had an unplanned reoperation, and 6 expired (Figure 1).
Table 1
| Variables | All (n=55) | RVPAS (n=17) | BTS (n=38) | P value |
|---|---|---|---|---|
| Age at Norwood procedure (days) | 5.0 (4.0–8.0) | 5.0 (4.0–6.0) | 6.0 (3.3–9.8) | 0.1 |
| Gestational age (weeks) | 38.6 (37.9–39.1) | 38.6 (38.0–39.0) | 38.7 (37.9–39.1) | 0.37 |
| Postmenstrual age at Norwood procedure (weeks) | 39.6 (38.7–40.0) | 39.4 (38.4–39.7) | 39.6 (38.9–40.0) | 0.14 |
| Weight at Norwood procedure (kg) | 3.0 (2.6–3.3) | 2.8 (2.6–3.2) | 3.1 (2.8–3.3) | 0.15 |
| ICU length of stay (days) | 12.0 (8.0–21.5) | 13.0 (8.0–30.0) | 11.0 (8.0–19.0) | 0.33 |
| Sex | 0.22 | |||
| Male | 31 (56.4) | 7 (41.2) | 24 (63.2) | |
| Female | 24 (43.6) | 10 (58.8) | 14 (36.8) | |
| Anatomy | 0.39 | |||
| HLHS | 45 (81.8) | 13 (76.5) | 32 (84.2) | |
| Unbalanced AVSD | 4 (7.3) | 2 (11.8) | 2 (5.3) | |
| Other | 6 (10.9) | 2 (11.8) | 4 (10.5) | |
| HLHS sub-type | 0.28 | |||
| MS/AS | 15 (33.3) | 6 (35.3) | 9 (23.6) | |
| MS/AA | 9 (20.0) | 4 (23.5) | 5 (13.2) | |
| MA/AA | 18 (40.0) | 2 (11.8) | 16 (42.1) | |
| MA/AS | 3 (6.7) | 1 (5.9) | 2 (5.3) | |
| Chromosomal abnormalities syndromes | 0.14 | |||
| CHARGE syndrome | 1 (1.8) | 1 (5.9) | 0 | |
| DiGeorge syndrome | 1 (1.8) | 1 (5.9) | 0 | |
| Other chromosomal abnormalities | 7 (12.7) | 3 (17.6) | 4 (10.5) | |
| Required ECMO immediately post-operatively | 3 (5.4) | 0 (0.0) | 3 (7.9) | 0.58 |
| Cardiac arrest | 6 (10.9) | 1 (5.9) | 5 (13.1) | 0.74 |
| Disposition from ICU | 0.15 | |||
| To stepdown cardiac unit | 43 (78.2) | 14 (82.4) | 29 (76.3) | |
| Deceased | 6 (10.9) | 3 (17.6) | 3 (7.9) | |
| Return to OR | 6 (10.9) | 0 | 6 (15.8) |
Median (25th–75th percentiles) are reported for continuous variables and a two-sided Wilcoxon rank-sum test is used to compare two populations. Frequencies (percentages) are reported for categorical variables and a Chi-square test is used to test their independence in the contingency table. RVPAS, right ventricle to pulmonary artery shunt; BTS, Blalock-Taussig shunt; ICU, intensive care unit; HLHS, hypoplastic left heart syndrome; AVSD, atrioventricular septal defect; MS, mitral stenosis; AS, aortic stenosis; AA, aortic atresia; MA, mitral atresia; ECMO, extracorporeal membrane oxygenation; OR, operating room.
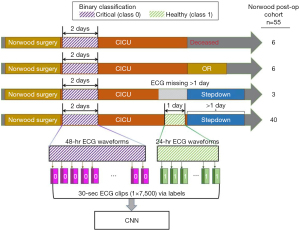
Data description
Continuous waveform data from ECG leads I, II, and III were collected from the ICU bedside monitors using the BedMaster system (Excel Medical Electronics, Jupiter, FL, USA), which is a third-party software connected to the hospital’s Philips monitors. There were 24 patients who had waveforms recorded at 125 Hz (prior to October 2017), which were resampled using interpolation to 250 Hz (14), and 31 patients were recorded at 250 Hz. Average missing data in leads I, II, and III per patient are 30.2%, 23.1%, and 35.0% respectively. Details regarding data quality are summarized in Table 2. Heart rate (HR), mean blood pressure (MBP), diastolic blood pressure (DBP), systolic blood pressure (SBP), and SpO2 were sampled from the BedMaster system once per second, whereas blood pressure data was first sampled from the arterial line data when available and then from non-invasive blood pressure values (i.e., after the arterial line removed). Laboratory values of pH, lactic acid, and base deficit were acquired from the electronic medical record (EMR) database.
Table 2
| Variables | Training cohort (n=45) | Test cohort (n=10) |
|---|---|---|
| Percentage of missingness in ECGs, mean (min, max) | 45.1 (2.4, 100.0) | 2.8 (2.5, 3.2) |
| Lead I | 27.2 (1.9, 91.8) | 2.7 (2.5, 3.2) |
| Lead II | 51.3 (2.4, 100.0) | 10.8 (2.5, 44.9) |
| Lead III | 45.1 (2.4, 100.0) | 2.8 (2.5, 3.2) |
| Number of non-empty samples, n (% labeled as 1) | ||
| Lead-I 30-sec clips | 162,692 (24.7) | 71,342 (26.6) |
| Lead-II 30-sec clips | 224,217 (22.6) | 71,364 (26.7) |
| Lead-III 30-sec clips | 124,224 (35.0) | 60,785 (31.3) |
ECG, electrocardiogram; min, minimum; max, maximum; Lead, electrocardiogram lead; sec, second.
Data preparation
The patient data was observed as a binary classification task where critically ill was defined as class 0 and stable as class 1. For model training, the patient data in the first two postoperative days were class 0 and the last day prior to transfer out of the ICU was class 1. Each lead’s waveform data was segmented into 30-second clips, resulting in vectors that are labeled as either a 0 or 1 depending on the day of the sample as described (Figure 1). The data was split into a training cohort (n=45) and test cohort (n=10). Makeup of the cohorts can be seen in Table S1. To ensure robust data, the first and last 30 minutes of data were omitted from the analysis to eliminate periods where the patient was being changed to different monitoring equipment. Data clips were removed where the entire 30 seconds was not captured (i.e., missing values) from that lead but clips from other leads from the same time period were retained. Waveform missingness and total included samples are summarized in Table 2. In total, the length of ECG signals for training was more than 15 million seconds and contained over 3.8 billion numerical values. It forms a sufficiently large training set of 170 thousand samples (i.e., 30-second ECG clips) per each lead for model fitting of CNNs.
Raw ECG clips were zero padded if any values were missing. Vital signs and laboratory values, which are non-continuously recorded, were aligned with continuous waveform data and each instance was set to the most recently recorded intermittent vital sign or laboratory value. Each intermittent value at each point in time was associated with a variable to indicate whether the value was inherited or new. All data were transformed through minimum-maximum normalization with the minimum and maximum values computed globally per each feature in the training set (15).
Algorithm development and training
The 10-layer CNN designed for use in this study is based on ResNeXt architecture which is a state-of-the-art CNN derived from ResNet (16,17). Its architecture was adapted for one-dimensional (1D) physiological streams inputs, rather than the two-dimensional (2D) streams, as originally designed. To accomplish this, the kernel in the convolutional layer (Conv) was modified to be a 1D stripe rather than a 2D patch and the model was trained de novo rather than utilizing existing, pre-trained models. The CNN was trained on 30-second ECG clips separately for each lead such that binary cross-entropy loss in the training data was minimized. The CNN layers contain a Conv, 4 aggregated residual blocks, a densely connected layer, and the final softmax activation. An aggregated residual block contains 32 paths, where input tensors are divided into 32 channels, each running through two successive convolutional and normalization layers and are concatenated together at the end.
The structure of the developed CNN is presented in Figure 2. In detail, the kernel size in each Conv is set to 16; the number of kernels is set to 64 in the first Conv layer and then identical in the residual blocks. Inputs are down-sampled by a factor of two at every two blocks by setting stride to 2 in Conv layer and max pooled (Pool) for the skip connections. To improve the training process, the normalization layer was set as a combination of batch normalization (BN), rectified linear unit (ReLU), and dropout (DO), so called BN-ReLU-DO normalization (18-20). DO rates were set to 0.5. The output, or final predictions, were made by a fully connected dense layer and softmax activation.
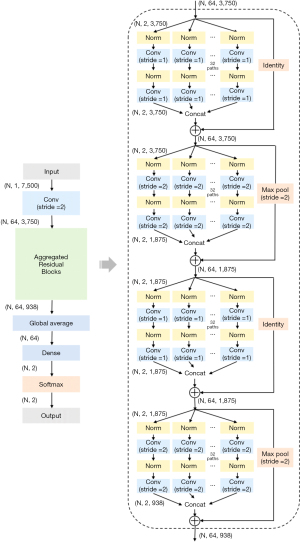
Adam optimizers with back-propagation were used for training the CNN developed as above (21). Learning rate was initially set to 10−3, and then reduced by a factor of 10−1 if the training loss has stopped decreasing. Batch size was set to 28. We tuned hyperparameters such as the kernel size in each Conv between {8, 16, 32, 64, 128} and the number of residual blocks between {2, 4, 8, 16}, by randomly selecting 85% of the training cohort for training and the rest 15% data for validation. Number of epochs in training was initially set to 20, but was stopped early when validation loss started increasing. By picking the model having the least validation error, we chose a kernel size of 16 and 4 residual blocks in our final model, as presented in Figure 2. We used 3-fold cross-validation to grid search the hyperparameters of traditional ML algorithms [i.e., L-1 regularizer in LR, depth in decision tree (DT), and number of trees in RF].
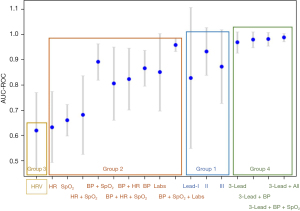
Definition of method groups for comparison
This study compared four methods of training AI and ML for their predictive value. Group 1 is the CNN model developed for this study and was trained on waveforms from ECG leads I, II, III, or a unified model combining all three. Group 2 utilized traditional ML algorithms and were trained on discrete vital sign and lab result data for the same binary classification task as the CNN. Group 3 utilized the same traditional ML algorithms and were trained based on 15 HR variability features, extracted from the lead-II ECG waveforms utilizing toolbox BioSPPy 0.6.1 in Python 3 (22). Group 4 combined the CNN models from Group 1 with the traditional ML algorithms from Group 2 by averaging their predictions.
Due to the much lower sampling frequency of vital sign and laboratory data, compared with waveform data, the data windows for the traditional ML algorithms applied to vital signs had to be longer. To yield a length of 300 points (1 vital sign sample per second), a window length of 5 minutes was used. Ten predictions, based on 30-second data windows, were averaged from Group 1, the CNN group, to yield a 5-minute window comparable to the traditional ML algorithms in Groups 2, 3, and 4.
Comparison to retrospective clinical observations
The models developed in this study compute time-varying scores which are the predicted probabilities of becoming stable (class 1) based on the observed trajectory of the patient’s changes of status (i.e., class 0—critical, class 1—stable). For each of the ten patients in the test cohort, the patient’s clinical stability was scored by a physician who was blinded to the results of the automated analysis. The score was recorded by retrospective chart review every 2 hours for the duration of the ICU stay into one of four categories: critical [1], moderate illness [2], mild illness [3], or stable [4]. Patients who were intubated, on more than one inotrope, require continuous anti-hypertensive agents, or had a lactate ≥3 mmol/L were classified as critical illness. Patients who were extubated on non-invasive positive pressure ventilation or high flow nasal cannula, on one inotrope, or had mild acidosis with lactate 2–3 mmol/L were classified as moderate illness. Patients who were extubated on high flow nasal cannula >3 L/minute, on no inotropic support, tolerated some feeds, and with lactate <2 mmol/L were classified as mild illnesses. Patients who were on no continuous infusions, on nasal cannula, on high flow nasal cannula <3 L/minute, and tolerating full feeding were classified as healthy. The study team developed these scores and are considered institutional norms for categorizing these patients.
Statistical analysis
All statistical analyses were performed using package SciPy 1.4.0 in Python 3 (14). For all continuous variables we report median (25th–75th percentiles) and utilize a two-sided Wilcoxon rank-sum test while comparing two populations. The predictive value of each AI algorithm was assessed utilizing the area under the curve (AUC) of the receiver operating characteristic (ROC) and the AUC of precision recall (PR) on each algorithm’s predictions of critical or stable (i.e., negative class 0 or positive class 1) for each patient in the test cohort (23). Precision calculates the percentage of positive predictions that were truly positive, whereas recall calculates the percentage of true positives that were correctly identified by the model. The scores for each patient were computed and presented in aggregate as mean (± standard deviation).
Results
The average ICU length of stay (LOS) in this study was 18.6 days (range, 7–85 days). A total of 43 patients (78.2%) were successfully discharged from the ICU to a general care cardiac telemetry unit, 6 (10.9%) expired, and 6 (10.9%) returned for re-operation (Figure 1).
Performance of the novel CNN and ML algorithms on the test cohort
Detailed performance data is presented in Table 3. For Group 1, the CNN deep learning model AUC-ROC values on the test cohort were 0.83 (±0.28), 0.93 (±0.09), and 0.87 (±0.15) based on data from ECG leads I, II, and III respectively. For Group 2, RF outperformed LR and DT with all non-waveform data, except laboratory data, where LR was superior. The best performing model in Group 2 was blood pressure, SpO2, and laboratory data taken together by averaging predictions from RF and LR which achieved an AUC-ROC of 0.95 (±0.03). For Group 3, the AUC-ROC was 0.62 (±0.15) utilizing RF on the HR variability data. For Group 4, the combination of CNN applied to ECG three-lead waveform data, RF applied to discrete vital sign data, and LR applied to lab results performed the best. This model ensemble approach naturally takes in different input modalities at different paces and with different missingness, and it often outperforms single models in predictions. Overall, this ensemble approach yielded an AUC-ROC of 0.98 (±0.02) and AUC-PR of 0.97 (±0.04). The AUC-ROC results are summarized graphically in Figure 3.
Table 3
| Group | Method | AUC-ROC | AUC-PR | Accuracy | Precision | Recall |
|---|---|---|---|---|---|---|
| 1 | Lead I | 0.83 (±0.28) | 0.79 (±0.26) | 0.36 (±0.07) | 0.34 (±0.03) | 0.99 (±0.01) |
| Lead II | 0.93 (±0.09) | 0.82 (±0.18) | 0.86 (±0.14) | 0.74 (±0.121) | 0.87 (±0.27) | |
| Lead III | 0.87 (±0.15) | 0.87 (±0.16) | 0.77 (±0.12) | 0.49 (±0.49) | 0.30 (±0.38) | |
| 2 | HR | 0.63 (±0.14) | 0.45 (±0.16) | 0.61 (±0.09) | 0.43 (±0.17) | 0.42 (±0.18) |
| SpO2 | 0.66 (±0.06) | 0.45 (±0.06) | 0.64 (±0.03) | 0.45 (±0.07) | 0.31 (±0.08) | |
| HR + SpO2 | 0.68 (±0.16) | 0.52 (±0.20) | 0.68 (±0.09) | 0.49 (±0.23) | 0.34 (±0.22) | |
| BP + HR + SpO2 | 0.81 (±0.14) | 0.71 (±0.20) | 0.75 (±0.10) | 0.74 (±0.19) | 0.45 (±0.25) | |
| Labs | 0.85 (±0.15) | 0.61 (±0.30) | 0.69 (±0.12) | 0.36 (±0.29) | 0.57 (±0.46) | |
| BP + HR | 0.82 (±0.12) | 0.73 (±0.17) | 0.75 (±0.09) | 0.74 (±0.16) | 0.48 (±0.23) | |
| BP | 0.87 (±0.07) | 0.81 (±0.10) | 0.82 (±0.06) | 0.87 (±0.12) | 0.55 (±0.14) | |
| BP + SpO2 | 0.89 (±0.07) | 0.82 (±0.12) | 0.84 (±0.07) | 0.89 (±0.07) | 0.58 (±0.17) | |
| BP + SpO2 + Labs | 0.95 (±0.03) | 0.92 (±0.05) | 0.85 (±0.09) | 0.92 (±0.07) | 0.63 (±0.32) | |
| 3 | HRV | 0.62 (±0.15) | 0.51 (±0.14) | 0.68 (±0.03) | 0.53 (±0.22) | 0.28 (±0.19) |
| 4 | 3-lead | 0.97 (±0.04) | 0.93 (±0.140 | 0.88 (±0.11) | 0.79 (±0.14) | 0.88 (±0.27) |
| 3-lead + BP | 0.98 (±0.03) | 0.95 (±0.07) | 0.93 (±0.08) | 0.91 (±0.10) | 0.88 (±0.27) | |
| 3-lead + BP + SpO2 | 0.98 (±0.03) | 0.95 (±0.08) | 0.92 (±0.09) | 0.91 (±0.10) | 0.89 (±0.24) | |
| 3-lead + BP + SpO2 + Labs | 0.98 (±0.02) | 0.97 (±0.04) | 0.92 (±0.09) | 0.93 (±0.08) | 0.85 (±0.30) |
Data are presented as mean (± standard deviation). AUC-ROC, area under the curve of the receiver operating characteristic; AUC-PR, area under the curve of precision recall; Lead, electrocardiogram lead; HR, heart rate; SpO2, peripheral blood oxygen saturation; BP, blood pressure; Labs, pH, lactic acid, and base deficit; HRV, heart rate variability.
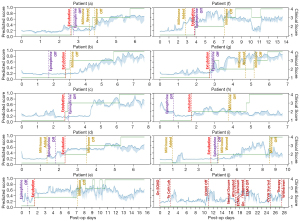
The ensemble model from Group 4 was performed on the test cohort to generate a value between 0 (critical) and 1 (stable), termed the Clinical Stability Score (CSS). Figure 4A demonstrates the CSS over the course of an ICU stay for patient (b) in the test cohort. In the test cohort, 7 of 9 patients (78%) who were transferred to the floor had a CSS of ≥0.5 at the time of transfer. Figure 4B is a 2D projection of the CNN-transformed ECG embeddings.
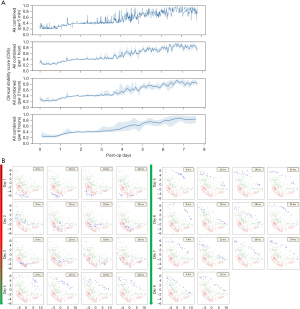
The CSS of the ten test patients was compared to the clinical observation score from retrospective chart reviews, where Spearman’s rank correlation was computed between the two ranked scores. They were closely correlated 78% of the time throughout their ICU stay (24). Figure 5 shows plots of the CSS and clinical observation scores over time for each of the ten patients in the test cohort.
Discussion
The results of this study demonstrate a novel application of AI in ICU medicine. The final model demonstrated precise discrimination between a critically ill patient and stable patient with an AUC-ROC of 0.98 (±0.02), accuracy of 92%, and precision of 93% (Table 3). Clinical assessment of neonates following the Norwood procedure is difficult and tools to help discriminate between a critically ill patient and stable patient are still needed. Such tools could prove invaluable in preventing critical events as well as predicting those patients who are ready to progress clinically. Monitoring patients more accurately for clinical progression could decrease ICU LOS and hospital LOS which would be impactful for patients, families, and the healthcare system.
Clinical prediction models have been developed for use in critical care to provide automated scores based on aggregated data in the EMR (25-27). One study, completed in 25 children after stage-1 surgical palliation for single ventricle heart disease, created an AI model optimized to detect impending clinical deterioration events (27). The AUC-ROC was 0.91 with good performance of the model noted 1–2 hours prior to the deterioration event. Another study in 1,445 pediatric ICU patients utilized seven vital signs as well as patient age and weight at ICU admission to develop a CNN predicting mortality 6–60 hours ahead of events (28). The AUC-ROC for prediction of mortality was 0.97 at 6 hours and 0.89 at 60 hours. The results of the present study achieved an AUC-ROC of 0.98 that focused on discriminating between clinical wellness and clinical instability, rather than focusing on detecting only deterioration events.
Close correlation between the CSS and the clinical observation score 78% of the time is interesting, and the periods where the two did not correlate merit further discussion as this is where the model may provide additional insights into clinical status. In Figure 5, patient (a) had an excellent correlation between the CSS and clinical observation score. Patient (a) demonstrated improvement in the CSS for 12–24 hours before inotropes were weaned or the patient was extubated. Though patient (e) did not correlate perfectly, the CSS was noted to decline at the time or just before the change in clinical observation score. Patient (e) had feeding intolerance intermittently that changed the clinical observation score but the CSS, which does not include feeding information, also demonstrated a decline in patient status at those times. Though the clinical observation score remained at 4 (i.e., ready to transfer), the patient was not transferred to stepdown until the CSS increased consistently to above 0.6, which suggests that some clinical uncertainty remained about this patient’s readiness for transfer that was captured by the CSS but not by clinical observation. Patient (i) was noted to have a clinical observation score of 0.4 at the time of transfer to stepdown, but the CSS had observed a decline in patient status, and this patient ultimately was readmitted to the ICU for heart failure six days after transferring out.
The goal, therefore, of adapting this AI model to a real-time assessment at the bedside would be to indicate wellness as a prompt to progress clinical care, as well as indicate deterioration as a prompt to investigate possible changes to clinical care that are needed. This represents a novel application compared to most existing clinical applications of AI.
Limitations
This model was designed for a very specific population of critically ill neonates in the cardiac ICU. Therefore, this model only applies to that population. Generalizing this model would require a broader training data set that would likely include additional variables such as primary cardiac diagnosis which is not needed in a model where the training set contains only one primary cardiac diagnosis. The developed model is able to recognize differences between the ECG waveforms from the critical and stable time periods, but this method does not provide any interpretable clinical insights in the waveforms due to the lack of interpretability in CNN models. The algorithm was trained on the data set from a single center and is subject to inherent bias from that organization. Certain elements of our patient population, surgical techniques, and critical care treatment protocols may affect performance of this same algorithm applied to patients from a different center. The retrospective clinical observation score is not validated by other studies and could be susceptible to center bias.
While the Group 4 combined algorithm performs well in the test cohort, prospective enrollment and monitoring is required to compare clinical evaluation at the moment to the CSS provided by the algorithm. Retrospective clinical observation score is likely biased by institutional practice and retrospective nature. Furthermore, bedside evaluation of how the tool performs in improving patient outcomes, decreasing ICU LOS, or decreasing hospital LOS will need to be prospectively validated. Future directions will include the development of a real-time tool as well as evaluation of additional factors that may be predictive of clinical wellness at the bedside that could not be incorporated in the current study (e.g., NIRS waveforms, continuous ventilator data).
Conclusions
This study demonstrates the successful development of an AI based algorithm utilizing ECG waveforms to differentiate between critically ill and stable patients following the Norwood operation. This model represents a novel application of AI in clinical medicine that extends beyond prediction of clinical deterioration, providing continuous assessment as patients progress from critical to stable. This work provides the basis for development of a real-time, bedside AI monitor to be prospectively validated, expanding the potential clinical applications of AI in medicine.
Acknowledgments
Funding: None.
Footnote
Reporting Checklist: The authors have completed the STARD reporting checklist. Available at https://jmai.amegroups.com/article/view/10.21037/jmai-22-35/rc
Data Sharing Statement: Available at https://jmai.amegroups.com/article/view/10.21037/jmai-22-35/dss
Peer Review File: Available at https://jmai.amegroups.com/article/view/10.21037/jmai-22-35/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://jmai.amegroups.com/article/view/10.21037/jmai-22-35/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by Children’s Healthcare of Atlanta (CHOA IRB #372) and Georgia Institute of Technology (GA Tech IRB #H18163) Institutional Review Boards and a waiver of informed consent was granted as all data would be retrospective and de-identified.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Ohye RG, Sleeper LA, Mahony L, et al. Comparison of shunt types in the Norwood procedure for single-ventricle lesions. N Engl J Med 2010;362:1980-92. [Crossref] [PubMed]
- McHugh KE, Pasquali SK, Hall MA, et al. Cost Variation Across Centers for the Norwood Operation. Ann Thorac Surg 2018;105:851-6. [Crossref] [PubMed]
- Theilen U, Shekerdemian L. The intensive care of infants with hypoplastic left heart syndrome. Arch Dis Child Fetal Neonatal Ed 2005;90:F97-102. [Crossref] [PubMed]
- Pickering BW, Herasevich V, Ahmed A, et al. Novel Representation of Clinical In-formation in the ICU: Developing User Interfaces which Reduce Information Over-load. Appl Clin Inform 2010;1:116-31. [Crossref] [PubMed]
- Manor-Shulman O, Beyene J, Frndova H, et al. Quantifying the volume of docu-mented clinical information in critical illness. J Crit Care 2008;23:245-50. [Crossref] [PubMed]
- Bergl PA, Nanchal RS, Singh H. Diagnostic Error in the Critically III: Defining the Problem and Exploring Next Steps to Advance Intensive Care Unit Safety. Ann Am Thorac Soc 2018;15:903-7. [Crossref] [PubMed]
- Marquet K, Claes N, De Troy E, et al. One fourth of unplanned transfers to a higher level of care are associated with a highly preventable adverse event: a patient record review in six Belgian hospitals. Crit Care Med 2015;43:1053-61. [Crossref] [PubMed]
- Croskerry P. Achieving quality in clinical decision making: cognitive strategies and detection of bias. Acad Emerg Med 2002;9:1184-204. [Crossref] [PubMed]
- Custer JW, Winters BD, Goode V, et al. Diagnostic errors in the pediatric and neo-natal ICU: a systematic review. Pediatr Crit Care Med 2015;16:29-36. [Crossref] [PubMed]
- Buch VH, Ahmed I, Maruthappu M. Artificial intelligence in medicine: current trends and future possibilities. Br J Gen Pract 2018;68:143-4. [Crossref] [PubMed]
- Davenport T, Kalakota R. The potential for artificial intelligence in healthcare. Future Healthc J 2019;6:94-8. [Crossref] [PubMed]
- Olive MK, Owens GE. Current monitoring and innovative predictive modeling to improve care in the pediatric cardiac intensive care unit. Transl Pediatr 2018;7:120-8. [Crossref] [PubMed]
- Yamashita R, Nishio M, Do RKG, et al. Convolutional neural networks: an overview and application in radiology. Insights Imaging 2018;9:611-29. [Crossref] [PubMed]
- SciPy community. SciPy 1.4.0 Release Notes. Updated June 4, 2020. [Accessed June 4, 2020]. Available online: https://scipy.github.io/devdocs/release.1.4.0.html
- Pedregosa F, Varoquaux G, Gramfort A, et al. Scikit-learn: Machine Learning in Python. J Mach Learn Res 2011;12:2825-30.
- Xie S, Girshick RB, Dollár P, et al. Aggregated Residual Transformations for Deep Neural Networks. 2017 IEEE Conference on Computer Vision and Pattern Recogni-tion (CVPR); 21-26 July 2017; Honolulu, HI, USA. IEEE; 2017:5987-95.
- He K, Zhang X, Ren S, et al. Deep residual learning for image recognition. 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR); 27-30 June 2016; Las Vegas, NV, USA. IEEE; 2016:770-8.
- He K, Zhang X, Ren S, et al. Identity Mappings in Deep Residual Networks. In: Leibe B, Matas J, Sebe N, et al. editors. Computer Vision – ECCV 2016. Lecture Notes in Computer Science, vol 9908. Springer, Cham; 2016:630-45.
- Ioffe S, Szegedy C. Batch Normalization: Accelerating Deep Network Training by Reducing Internal Covariate Shift. ICML'15: Proceedings of the 32nd International Conference on International Conference on Machine Learning. 2015;37:448-56.
- Srivasava N, Hinton G, Krizhevsky A, et al. Dropout: a simple way to prevent neural networks from overfitting. J Mach Learn Res 2014;15:1929-58.
- Kingma DP, Ba J. Adam: A Method for Stochastic Optimization. arXiv:1412.6980. [Preprint]. 2015. Available online: https://doi.org/
10.48550/arXiv.1412.6980 - BioSPPy. BioSPPy v0.6.1 API Reference. Updated August 29, 2017. [Accessed June 4, 2020]. Available online: https://biosppy.readthedocs.io/en/v0.6.1/biosppy.html
- Powers D. Evaluation: From Precision, Recall and F-Measure to ROC, Informedness, Markedness & Correlation. Journal of Machine Learning Technologies 2011;2:37-63.
- Spearman Rank Correlation Coefficient. In: The Concise Encyclopedia of Statistics. New York, NY: Springer, 2008. Available online:
10.1007/978-0-387-32833-1_379 10.1007/978-0-387-32833-1_379 - Pollack MM, Patel KM, Ruttimann UE. The Pediatric Risk of Mortality III--Acute Physiology Score (PRISM III-APS): a method of assessing physiologic instability for pediatric intensive care unit patients. J Pediatr 1997;131:575-81. [Crossref] [PubMed]
- Beltempo M, Shah PS, Ye XY, et al. SNAP-II for prediction of mortality and mor-bidity in extremely preterm infants. J Matern Fetal Neonatal Med 2019;32:2694-701. [Crossref] [PubMed]
- Rusin CG, Acosta SI, Shekerdemian LS, et al. Prediction of imminent, severe dete-rioration of children with parallel circulations using real-time processing of physio-logic data. J Thorac Cardiovasc Surg 2016;152:171-7. [Crossref] [PubMed]
- Kim SY, Kim S, Cho J, et al. A deep learning model for real-time mortality predic-tion in critically ill children. Crit Care 2019;23:279. [Crossref] [PubMed]
Cite this article as: Aljiffry A, Xu Y, Hong S, Long JB, Sun J, Maher KO. Artificial intelligence and clinical stability after the Norwood operation. J Med Artif Intell 2023;6:26.


