Using a machine learning model to risk stratify for the presence of significant liver disease in a primary care population
Highlight box
Key findings
• Machine learning may aid strategic tackling of the diagnostic challenge of identification of presence of early liver disease at a population level.
• An Ensemble Stacker model showed the best performance at classifying a patient as at high risk or low risk of liver disease, with the performance shown by an area under the curve (AUC) of 0.72, calculated within an unseen hold-out data set.
What is known and what is new?
• There is significant heterogeneity between techniques used to date to develop machine learning models for diagnosis and risk stratification for liver disease.
• To the authors knowledge there is no other training cohort solely derived from a primary care population; this reinforces likely translatability of our work.
What is the implication, and what should change now?
• Machine learning approaches for development of a potential screening tool for liver disease in the community need to be explored with comparison to current screening tools undertaken.
Introduction
Background
Globally, chronic liver disease (CLD) is an emerging epidemic with challenges in diagnostics and clinical management at both a patient and population level. Any chronic injury results in liver fibrosis (scarring), which is reversible in the initial stages due to the regenerative capacity of the liver. However, progression from fibrosis to advanced cirrhosis can result in liver failure which is associated with high risk of mortality for patients and a high healthcare burden for society. Liver disease has been dubbed ‘the silent killer’ as often there are only symptoms in late stages of disease; approximately 50% of patients are first diagnosed with liver disease on an emergency admission to hospital (1).
Lifestyle related liver damage is the most common cause worldwide of liver disease with non-alcoholic fatty liver disease (NAFLD) being present in approximately 25% of the population globally (2). Combined with an aging population, an exponential rise in liver disease due to increasing obesity and type 2 diabetes mellitus (T2DM), is expected (3). In Europe, alcohol liver disease (ALD) is also increasing and attributed to aetiology of disease in over 50% of cirrhotic patients. Moreover, ALD is the 2nd highest cause of years of working life lost in the UK (4,5). There are no licensed treatments for lifestyle related liver disease and internationally there is a call by experts for the need for fundamental change in order to prevent disease; both by decreasing influential lifestyle factors and improving early disease diagnosis when reversibility of the condition is possible (5).
Rationale and knowledge gap
Effectively testing for liver disease is challenging because symptoms occur late. The reliance on liver functions tests (LFTs), which has been the traditional method of diagnosing suspected CLD, is flawed. LFTs often do not detect significant liver disease; up to 90% of patients with severe liver disease have normal LFTs (6). In contrast, up to 20% of LFTs are abnormal, which leads to possible over investigation as the majority of these patients never develop liver disease (7). Non-invasive diagnostic tests such as transient elastography (TE) and the serum enhanced liver fibrosis (ELF) tests are only available in certain geographical areas and are often relatively expensive and largely based in secondary care (8). There are a number of further challenges to current diagnostics in CLD within the context of a community setting. Firstly, many of these tests have been derived in secondary care populations and then extrapolated to a community setting. The prevalence of disease and spectrum bias will differ between secondary care and community care, thus questioning the validity of this extrapolation. Secondly, when routine blood tests are used to derive diagnostic algorithms using traditional statistical techniques, they have been found to have limitations. For example, a score called Fibrosis-4 (FIB4), which contains the liver enzyme tests aspartate aminotransferase (AST), alanine transaminase (ALT) and platelet count, is used in clinical pathways to determine the need for further investigation, but has been shown to miss people with significant liver disease (9). Finally, many of the tests are disease specific but there is increasing awareness of dual pathology, e.g., alcohol and metabolic related liver disease, and thus having a diagnostic strategy that attends to this issue is critical.
Machine learning (ML) is a novel approach in this area; conventional statistical methods previously used may not have captured the non-linear relationships between biochemical and demographic variables especially when there are phenotypically different sub-populations of lifestyle related disease i.e., NAFLD vs. ALD. Models could match healthcare provision with patient needs, which could save resources in comparison to the current ‘one size fits all’ approach to a populations’ patients who have different individual characteristics (10).
ML techniques therefore provide a novel way of identifying significant variables without pre-conceived bias. As a proof-of-concept study, we wanted to examine the performance of 9 different ML models in a large community cohort based on the presence of abnormal LFTs and/or risk factors for alcohol and metabolic related liver disease.
Objective
To create a diagnostic algorithm using ML to predict the presence of clinically significant liver fibrosis using demographics and routine investigation results available in primary care. TE will be used as the ground truth as the surrogate marker of the presence or absence of liver fibrosis. For maximum clinical relevance, these results will be extrapolated to state if the patient has either low risk of liver disease or high risk of having underlying CLD indicating whether further investigation is required for liver disease beyond basic tests. We present this article in accordance with the STARD reporting checklist (available at https://jmai.amegroups.com/article/view/10.21037/jmai-23-35/rc).
Methods
Materials
Data collection
Over 25,000 patients from five different general practitioner (GP) practices were identified as part of a research study to investigate early diagnosis of liver disease in a community setting. All patients were located within areas of different socioeconomic status in Nottinghamshire and Leicestershire (UK) and were screened for risk factors for CLD (6,11). Risk factors for liver disease included a diagnosis of T2DM, obesity, abnormal liver blood tests and a history of above recommended alcohol consumption and were identified by NHS Read codes as previously published (6,12). Patients over 18 years old who met inclusion criteria were invited to attend a clinic appointment in which routine blood tests and TE were carried out. Routine demographic data, investigation results and medical history were documented, and these will be referred to as model variables. Patients already known to have CLD or be under the care of the hospital liver team were excluded. A total of 1,453 patients were consecutively prospectively recruited. For these patients, other specific liver disease pathologies were ruled out via tests and clinical assessment. For this proof of concept study the patients were recruited within a set time frame.
TE was carried out using Fibroscan (Echosens), which gave a numerical indication in kilopascals (kPa) of liver stiffness, and was used as the ground truth as the test is widely validated for use in diagnosis and screening for liver fibrosis in the community or out-patient setting (8). Internationally, a threshold of 8 kPa is applied, below which clinically significant liver fibrosis is excluded and above which more extensive investigation is warranted (8,13). There were no reported adverse events.
Ethics
This data collection was carried out from 2012–2016 as part of the ongoing project of ‘Stratification of Liver Disease in the Community Using Fibrosis Biomarkers’ project as approved by the Leicester Research Ethics Committee (13/EM/0123) with the clinicaltrials.gov identifier: NCT02037867. This was used as the evidence base in developing the commissioned pathway starting in 2016 which has been previously reported (9). The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). Informed consent was taken from all the patients. Informed consent was taken from all the patients.
Dataset formation
During the initial study, three geographical locations were visited sequentially over the 4-year study period. The chronologically recruited patients were then split into two datasets with 2/3 of total patients forming the ‘Training dataset’ and a 1/3 the ‘Hold out dataset’ as shown in Figure 1.
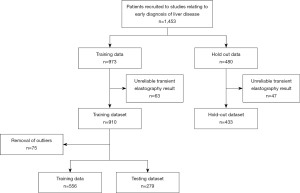
To avoid data leakage the initial ‘Training dataset’ was split at the beginning of the pipeline into ‘Training data’ and ‘Testing dataset’ with a 70%/30% split respectively and an equal number of patients with high TE reading in each dataset (15). This contrasts with the normal practice of processing the training and testing datasets through the same pipeline, which can lead to the model being biased towards the testing dataset and may, in turn, lead to the overfitting of a developed algorithm. The algorithm developers had access to the TE results for the ‘Training dataset’ as this was the ground truth used for algorithm training. Only the clinical team had access to the TE results for the ‘Hold out set’ to provide unseen data for final algorithm validation.
Methods: algorithm development
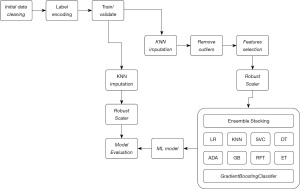
An overview of the ML learning pipeline constructed to build the ML model is outlined in Figure 2.
Data discovery and algorithm development
Data correlations
To investigate the relationship between different variables and TE, correlations were carried out using Pearson correlation coefficient. Significant correlations shown included TE with AST, alkaline phosphatase (ALP), ALT, and body mass index (BMI) with coefficients 0.33, 0.21, 0.22 and 0.25 respectively as shown in Figure S1. Significant correlation between model features was not seen therefore no variables were removed based on this analysis from further steps.
ML classifier development
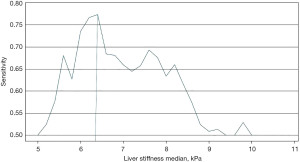
A threshold of a TE above 8 kPa is widely used to stratify patients who are likely to have clinically significant liver fibrosis. The use of a cut-off for the dataset training was explored in order to develop the ML classifier. As indicated in Figure 3 it was found that the model performed best when trained with a TE cut off at 6.2 kPa which is likely due to the more balanced dataset between those with ground truth indicating clinically significant liver fibrosis vs. those with low TE results. This is a well-recognised issue within ML with an imbalanced dataset leading to bias prediction towards the majority class (16). An imbalance graph is shown in Figure S2 indicates minimal imbalance in the dataset used.
When building the ensemble algorithm, a grid search was used to fine tune the hyperparameter’s performance. GradientBoostingClassifier, the meta learner, was set up using several hyperparameters. Estimators were set to n=1,000 and due to this being a classification task the loss function was configured as exponential. The maximum number of variables used was 6 to ensure individual models were built using a diverse set of variables. To overcome overfitting the maximum depth of each tree was 3. To introduce randomness and reduce the correlation between models a subsample ratio of 0.5 was used with a learning rate of 0.001. A random state was applied in order to ensure result reproducibility.
To train the model, the StratifiedKFold technique was carried out using the testing dataset. StratifiedKFold is a variant of k-fold cross-validation that ensures the preservation of the class distribution in each fold and was used due to good performance in imbalanced datasets. By using StratifiedKFold, the dataset was divided into k equal-sized folds while maintaining the same class distribution as the original data. During training, the model was trained and evaluated k times, with each fold serving as the validation set once while the remaining folds were used for training. This approach helps to mitigate the risk of overfitting and provides a more reliable estimate of the model’s performance on unseen data.
Missing data patterns in the training set
The dataset did not suffer from significant missing values (see Figure S3) with the variable with most missing values being HbA1c. This was expected as it is less frequently carried out within the non-diabetic population, with 14% and 0.5% of the non-diabetic and T2DM populations respectively not having the value recorded. Missing BMI values were shown to be missing at random.
The k-nearest neighbor (KNN) method was applied to impute missing values with K=3 as selected using the elbow method. KNN is a recognised to contribute significantly to classification performance (17).
Scaling variable results
The variables used within the dataset have multiple ranges and units due to the measured variables’ diverse nature which could have potentially led to a risk of data skewing. Scaling standardisation was carried out on all data using a z score method with RobustScaler. This used the interquartile range so that it was robust to outliers making the centre of attention on the bulk of the data (18,19).
Variable engineering (medicine and comorbidities)
Included in the dataset were open-text variables for each patient such as current medications and comorbidities. Pre-processing of the medication and comorbidities included elimination of noise and irrelevant information by removing specific recurrent or obsolete characters and then using WordNetLemmatizer to reduce words to base form. For comorbidities comma separation and generating n-grams of length 2 was carried out prior to running data through an International Classification of Disease 10th Revision (ICD-10) application programming interface (API) to retrieve the parent code (20). Patients were then assigned a positive classification if the comorbidity was present. Cosine similarity was used to compare medications with a pre-formulated list based on the British National Formulary (BNF) and assign a label for the parent class of medication. This was then reviewed by the clinical team to ensure correct categorisation.
Remove outliers
Identifying outliers in the data was essential to minimise potential confusion and bias. Tukey’s method for identifying outliers was carried out (21). Using this method, a total number of 75 patients have been removed. A breakdown of outliers detected and removed are shown in Figure S4.
Statistical analysis
Comparative analysis of cohort characteristics
Table 1 presents a summary of the key findings from our analysis of the training and hold-out patient population. We employed the t-test as a statistical hypothesis test to assess if there was a significant difference between the means of two groups for age and BMI variables. Chi-squared was employed to describe the gender, ethnicity, number of patients with abnormal liver blood tests and percentage of the population diagnosed with diabetes between the two datasets. Wilcoxon rank test was used to describe the TE and current alcohol use.
Table 1
| Dataset characteristic | Whole cohort (n=1,453) | Training (n=973) | Hold-out (n=480) | P value |
|---|---|---|---|---|
| Mean age (years) | 59 | 59 | 59 | 0.71 |
| Gender (% female) | 38.6 | 37.1 | 41.7 | 0.09 |
| Median transient elastography (kPa) | 5.5 | 5.6 | 5.4 | 0.02 |
| Mean BMI (kg/m2) | 29.7 | 28.6 | 32.1 | <0.05 (2.20×10−16) |
| Transient elastography greater than 8 kPa (%) | 20.3 | 22.2 | 16.5 | <0.05 (0.01) |
| T2DM (%) | 49.2 | 58.0 | 31.5 | <0.05 (7.44×10−62) |
| Current alcohol (median units/week) | 4 | 4 | 6 | 0.58 |
| Ethnicity (% Caucasian) | 76.9 | 73.4 | 84.2 | <0.5 (6.85×10−7) |
| Abnormal liver blood tests (% abnormal) | 20.4 | 21.7 | 17.7 | 0.77 |
BMI, body mass index; T2DM, type 2 diabetes mellitus.
Comprehensive evaluation metrics for model performance
Throughout the paper we utilized a comprehensive set of evaluation metrics to gain deeper insights into the effectiveness of different classifiers. The metrics employed encompassed a diverse range of performance aspects, enabling a robust comparison across various models. The area under the curve (AUC) served as an indicator of the classifiers’ overall discriminatory power, providing a concise summary of their ability to distinguish between classes. Sensitivity gauged the models’ capability to correctly identify positive instances, while Specificity measured their proficiency in correctly identifying negative instances. Additionally, the F1 score, which balances both precision and recall, and the classification report offered a more detailed breakdown of the classifiers’ performance, facilitating a comprehensive evaluation of their strengths and weaknesses. By employing this diverse set of metrics, we obtained a comprehensive understanding of the model’s performance, aiding us in making informed decisions regarding the most suitable classifier for our specific application.
Algorithm development
Variable selection
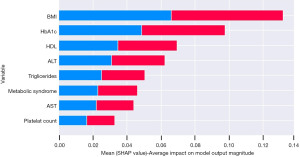
After variable engineering a total of 87 variables were used in the ML classifier development. As the number of variables increased, it enforced a new challenge to improve the ML classification (22). Performing variable selection techniques (i.e., recursive feature elimination) was used as a robust method to generate the best combination of variables relating to the ground truth (23). The importance of the variables was ranked by applying the RandomForest tree (RFE) and includes variables with a relative importance above 60 which is shown in Figure S5. A trade-off for the ML classifier was to reduce the number of variables to avoid redundancy whilst still achieving a performance above 60 and this was achieved. Overall, the most important variables were BMI, haemoglobin A1c (HbA1c), high-density lipoprotein (HDL), ALT, triglycerides, presence of metabolic syndrome, AST and platelet count.
Base line models
Table 2 shows how different ML models performed in the testing dataset. The testing dataset consisted of 274 patients with TE indicating no further investigation was required in 176 patients (TE <8 kPa) and further investigation was needed in 98 patients (>8 kPa). This has the same percentage of imbalance as in the full dataset. As the dataset suffered from an imbalance issue, the model output shows an improvement in the more complex ML algorithms (i.e., decision trees, bagging, stacking). The Ensemble stacking model and the Meta-learner Extra Tree Classifier show a good performance in the imbalanced dataset. The Ensemble stacking stands out for all reported criteria with a specificity of 69%, sensitivity of 83% and AUC of 0.75. Moreover, in the stratified KFold validation, the mean sensitivity is 73% and highest is 88%. The AUC is 0.74 (95% CI: 0.66–0.84).
Table 2
| Model | AUC | Sensitivity | Specificity |
|---|---|---|---|
| AdaBoost classifier | 0.50 (0.50–0.50) | 1 (1.00–1.00) | 0.00 (0.00–0.00) |
| CART | 0.66 (0.60–0.67) | 0.70 (0.58–0.70) | 0.68 (0.57–0.68) |
| Extra trees classifier | 0.68 (0.59–0.69) | 0.76 (0.62–0.76) | 0.64 (0.52–0.65) |
| KNeighbors classifier | 0.69 (0.60–0.69) | 0.72 (0.58–0.73) | 0.68 (0.56–0.68) |
| SVC | 0.70 (0.60–0.70) | 0.68 (0.53–0.68) | 0.76 (0.65–0.76) |
| Logistics regression | 0.71 (0.64–0.75) | 0.80 (0.68–0.82) | 0.66 (0.56–0.66) |
| Gradient boosting classifier | 0.73 (0.63–0.73) | 0.81 (0.67–0.82) | 0.68 (0.56–0.69) |
| Random forest classifier | 0.74 (0.65–0.74) | 0.76 (0.62–0.76) | 0.64 (0.52–0.65) |
| Ensemble stacking | 0.75 (0.63–0.75) | 0.83 (0.68–0.85) | 0.69 (0.56–0.70) |
AUC, area under the curve; CART, classification and regression tree; SVC, support vector classification.
Variable importance
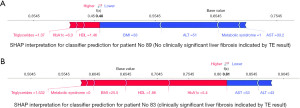
Using Shapley additive explanations (SHAP) to explain further the importance of the variables in the model, Figure 4 shows the relationship between each variable and the distribution of the impact of each variable. Figure 5 ‘Patient A’ shows the SHAP visualisation for how the classifier behaved in one patient where the output indicated the likely results was ≤8 kPa or ‘No’ further investigation required. As shown in blue the negative SHAP variables push the classification value in the negative direction towards an output of ‘No’. The SHAP values in red represent variables that push in the positive direction towards an output of ‘Yes’ for further investigation required. On the other hand, Figure 5 ‘Patient B’ shows a prediction where the classifier predicted that the TE result would be ≥8 kPa or that ‘Yes’—the patient would require further investigation for liver disease.
Results
Evaluation of model using hold-out data
The model performed with consistent results on the unseen hold out data; on testing using the testing dataset the ensemble stacking model produced an AUC of 0.75 in comparison to testing in the holdout data which yielded an AUC of 0.72 (Table 3). Table 4 compares the specificity and sensitivity of patients with different risk factors for liver disease. The method of analysing results and providing evidence of diagnostic accuracy was carried out as planned study inception.
Table 3
| Population | No. of patients | AUC | Sensitivity | Specificity |
|---|---|---|---|---|
| Hold out dataset | 433 | 0.72 (0.64–0.72) | 0.74 (0.65–0.81) | 0.60 (0.59–0.66) |
| Training cohort | 911 | 0.75 (0.63–0.75) | 0.83 (0.68–0.85) | 0.69 (0.56–0.70) |
AUC, area under the curve.
Table 4
| Hold-out data population | No. of patients | AUC | Sensitivity | Specificity |
|---|---|---|---|---|
| ≥2 comorbidities | 89 | 0.47 (0.35–0.59) | 0.54 (0.33–0.75) | 0.40 (0.32–0.48) |
| Alcohol + BMI + obesity | 26 | 0.58 (0.46–0.70) | 0.82 (0.66–1.00) | 0.34 (0.13–0.50) |
| T2DM only | 40 | 0.56 (0.56–0.73) | 0.92 (0.84–1.00) | 0.37 (0.22–0.47) |
| Obesity only | 180 | 0.59 (0.51–0.69) | 0.71 (0.56–0.88) | 0.48 (0.42–0.53) |
| Alcohol and obesity | 34 | 0.60 (0.42–0.79) | 0.66 (0.33–1.0) | 0.54 (0.41–0.67) |
| T2DM and obesity | 40 | 0.61 (0.54–0.70) | 0.91 (0.83–1.00) | 0.32 (0.19–0.44) |
| T2DM and alcohol | 31 | 0.64 (0.37–0.90) | 0.50 (0.00–1.00) | 0.78 (0.67–0.89) |
| ≥50 years old | 324 | 0.67 (0.62–0.72) | 0.69 (0.61–0.78) | 0.65 (0.62–0.69) |
| <50 years old | 109 | 0.78 (0.75–0.82) | 1.00 (1.00–1.00) | 0.57 (0.50–0.64) |
| Alcohol only | 76 | 0.84 (0.48–0.99) | 0.73 (0.00–1.00) | 0.94 (0.91–0.98) |
AUC, area under the curve; BMI, body mass index; T2DM, type 2 diabetes mellitus.
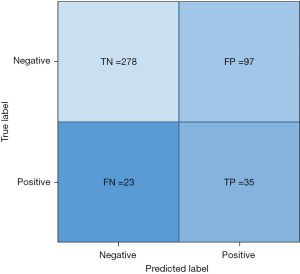
Further analysis of the performance of the model shown portrayed using the confusion matrix in Figure 6. Assuming a prevalence of clinically significant fibrosis in a high risk population as 22.9% as shown by Chalmers et al., the positive likelihood ratio is 1.85 with a negative likelihood ratio of 0.43 (9). The post-test probability of clinically significant liver disease for positive results is 35.46% and 11.33% for the negative results.
As part of exploratory analysis FIB-4 was carried out in the Hold-out dataset and an AUC of 0.51 was noted with a negative predictive value (NPV) of 0.85 vs. the Ensemble model’s AUC of 0.72 and NPV of 0.92.
Discussion
Key findings
This collaboration of clinicians and data scientists has shown that ML can be used to predict whether a patient in the community requires further investigation for liver disease. A parsimonious model using readily available variables has an AUC of 0.75 on internal testing and 0.72 when unseen hold out data was tested. The analysis highlights eight key variables including BMI, HbA1c, presence of metabolic syndrome, HDL, ALT, triglyceride, AST and platelet count results that underpin the model.
Strengths and limitations
A limitation of our study is the use of TE as the ground truth for indicating the presence of clinically significant liver disease rather than the gold standard test, a liver biopsy. Routine biopsy of patients with a low risk of liver disease, before non-invasive tests indicate presence of liver disease, would not be ethically correct or feasible. Furthermore, the cost of biopsy is approximately £500 with a 1% risk of complications (24,25). By using TE, the cohorts are more evenly weighted for patients with liver disease versus those without clinically significant liver disease. However TE can miss clinically significant liver disease if used as a standalone test; in development of LiverAID TE missed 9% of patients with F2-F4 fibrosis (26). Additionally there is approximately a 6% technical failure rate of Fibroscan which may minimally skew the training dataset (9). All patient with unreliable or failed scans were excluded from this study. The XL and M probe are used routinely in clinical practice within this clinical pathway which has been shown to increase the number of valid readings (27).
The model performs well across many of the sub-groups of disease which is important for the potential utility in a community setting with a wide range of risk factors. Some of the sub-groups are small, so care needs to be taken with interpretation of the data as the confidence intervals are large. The sub-group obesity (n=180) did appear to have a lower AUC than the overall group; AUC 0.59 (0.51–0.69). Due to the small cohort size of different geographical regions, we were unable to assess model performance in these individual groups. Further optimisation of the algorithm and validation in different subgroups and different geographical regions is needed.
It is interesting to note that the testing and hold out datasets had an equivalent performance despite having different patient characteristics. This reflects the variation of risk factors that exists in this study as it has been performed across different geographical areas. Thus, the stability of the overall model is encouraging and has implications for generalisability to other external cohorts.
Comparison with similar researches
Different ML models have used different ground truths and baselines populations for development. Much of the published work focuses on imaging and nonalcoholic steatohepatitis (NASH)/NAFLD diagnosis. Other ground truths used are ultrasonography, liver biopsy and TE with most datasets being derived from a post biopsy secondary care population.
Diagnostic models include NASHmap which used XGBoost, a technique using ‘gradient boosting’ in which there are iterative computations of weaker models, to develop both a 14 variable and 5 variable model using basic variables to predict a probable diagnosis of NASH using biopsy as a ground truth (28). A high performance of AUC =0.82 in predicting NASH was found when evaluated against a large claims derived database, Optum.
Perakakis et al. used 365 different lipid species, glycans and hormones measurements of 31 NAFLD patients to develop a ML algorithm to diagnose NAFLD without need for an invasive procedure (29). Using ten lipid species the accuracy of diagnosis of presence of liver disease was 98% but the extra panel of blood test cost $605.
A training set of over 10,000 patients was used by Liu et al. in eastern China to develop an algorithm aiming to diagnose NAFLD (30). All participants underwent either imaging or histological examination to determine presence or absence of NAFLD. Multiple ML techniques were trialed; XGBoost showed an AUC of 0.93 in the training set and an AUC of 0.87 in the validation set. BMI was shown as the most important indicator for presence of liver disease.
In Denmark the LiverAID models, ensemble learning classifiers, were trained using a pre-hospital cohort of 3,352 patients with 492 patients undergoing a liver biopsy (26). TE was used as the ground truth with 223 input variables gathered from prospectively collected data. Synthetic samples were generated to allow for class imbalance. The NPV when tested in a holdout set was over 96% for clinically significant liver disease in sub-cohorts of patients with ALD risk factors, NAFLD risk factors and patients approached from the general population. In this study the importance of a parsimonious end model was acknowledged; the aim is for a model which has adequate diagnostic capabilities but also uses least healthcare resource. When compared to the traditional non-invasive scores of FIB-4, Forns index and APRI, the LiverAID models performed well at predicting clinically significant liver disease as defined by a TE of >8 kPa (AUC 0.60–0.76 vs. 0.86–0.91, P=0.000–0.001).
The above models show the heterogeneity between training set derivation and methodology whilst highlighting the potential for improved patient identification and stratification. There is overall a need for external validation of developed ML algorithms and careful consideration of cost implications of sourcing incorporated variables.
Explanations of findings—the vision for clinical utility
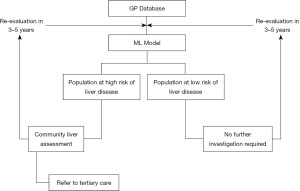
In today’s healthcare systems there are a number of potential uses for ML and exploration of this tool for population health management may help overcome limited resources and an increasing burden of disease. A benefit of all the non-invasive scoring systems including ML derived models is they are low risk to the patient as they use already collected variables and the procedures used to obtain these are relatively low risk, i.e., phlebotomy. An example of where the ID-LIVER algorithm could be applied is shown in Figure 7 with information feeding from a GP database to determine which patients need further assessment for liver disease. This could be carried out in a community setting, e.g., a Diagnostic Hub, which could offer specialist services in the patient’s neighbourhood.
Implications and actions needed
Clinical validation of this algorithm in an adult patient population who have risk factors for CLD is the next critical step. Whilst the diagnostic values of the ML algorithm are encouraging, an understanding of how the algorithm can be incorporated into clinical pathways is required. The Ensemble stacker’s superior performance to FIB-4 in this study at determining clinically significant fibrosis is supported by recently published data showing FIB-4’s effectivity as a screening tool in a population at high risk of liver disease need to be carefully considered (31). The focus of the algorithm could be adjusted depending on whether the question is about reducing the number of cases missed (enhancing NPV) or increasing the value of specialist tests [enhancing positive predictive value (PPV)]. The balance of this trade off needs careful thought and interaction with clinicians, patients and healthcare providers.
Conclusions
This paper is proof of concept that the use of ML should be explored to help strategic tackling of the diagnostic challenge of identification of early liver disease at a population level.
Acknowledgments
Funding: This article was supported by
Footnote
Reporting Checklist: The authors have completed the STARD reporting checklist. Available at https://jmai.amegroups.com/article/view/10.21037/jmai-23-35/rc
Data Sharing Statement: Available at https://jmai.amegroups.com/article/view/10.21037/jmai-23-35/dss
Peer Review File: Available at https://jmai.amegroups.com/article/view/10.21037/jmai-23-35/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://jmai.amegroups.com/article/view/10.21037/jmai-23-35/coif). L.B., M.M., R.H., H.P., M.P., V.S.A., K.P.H., N.A.H., and I.N.G. are supported by UKRI Innovate UK as part of ID-LIVER (project number 40896). L.B., J.R.M. and I.N.G. are supported by the Gastrointestinal and Liver Disorder theme of the NIHR Nottingham Biomedical Research Centre (Reference No. BRC-1215-20003). M.M., R.H., M.P. are current employees of Jiva.ai. K.P.H. is supported by the Medical Research Council (MRC; MR/P023541/1) and the Wellcome Trust (203128/Z/16/Z). J.R.M. is supported by the Medical Research Council Clinician Scientist award (MRC; MR/P008348/1), and has received investigator-led funding research from Gilead Sciences. Gilead had no intellectual input into the study concept or design, interpretation of the results or editing the manuscript. I.N.G. has received investigator-led funding research from Gilead Sciences. Gilead had no intellectual input into the study concept or design, interpretation of the results or editing the manuscript. V.S.A. has received funding from Norgine and Vertex. Norgine and Vertex have had no intellectual input into the study concept or design, interpretation of the results or editing the manuscript. The authors have no other conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by Leicester Research Ethics Committee, United Kingdom (13/EM/0123) and informed consent was taken from all the patients.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Ratib S, Fleming KM, Crooks CJ, et al. 1 and 5 year survival estimates for people with cirrhosis of the liver in England, 1998-2009: a large population study. J Hepatol 2014;60:282-9. [Crossref] [PubMed]
- Younossi Z, Anstee QM, Marietti M, et al. Global burden of NAFLD and NASH: trends, predictions, risk factors and prevention. Nat Rev Gastroenterol Hepatol 2018;15:11-20. [Crossref] [PubMed]
- Estes C, Razavi H, Loomba R, et al. Modeling the epidemic of nonalcoholic fatty liver disease demonstrates an exponential increase in burden of disease. Hepatology 2018;67:123-33. [Crossref] [PubMed]
- Poznyak V, Rekve D. editor. Global status report on alcohol and health 2018. World Health Organization; 2018.
- Karlsen TH, Sheron N, Zelber-Sagi S, et al. The EASL-Lancet Liver Commission: protecting the next generation of Europeans against liver disease complications and premature mortality. Lancet 2022;399:61-116. [Crossref] [PubMed]
- Harman DJ, Ryder SD, James MW, et al. Direct targeting of risk factors significantly increases the detection of liver cirrhosis in primary care: a cross-sectional diagnostic study utilising transient elastography. BMJ Open 2015;5:e007516. [Crossref] [PubMed]
- Dillon JF, Miller MH, Robinson EM, et al. Intelligent liver function testing (iLFT): A trial of automated diagnosis and staging of liver disease in primary care. J Hepatol 2019;71:699-706. [Crossref] [PubMed]
- European Association for the Study of the Liver. EASL Clinical Practice Guidelines on non-invasive tests for evaluation of liver disease severity and prognosis - 2021 update. J Hepatol 2021;75:659-89. [Crossref] [PubMed]
- Chalmers J, Wilkes E, Harris R, et al. The Development and Implementation of a Commissioned Pathway for the Identification and Stratification of Liver Disease in the Community. Frontline Gastroenterol 2020;11:86-92. [Crossref] [PubMed]
- Subramanian M, Wojtusciszyn A, Favre L, et al. Precision medicine in the era of artificial intelligence: implications in chronic disease management. J Transl Med 2020;18:472. [Crossref] [PubMed]
- Harman DJ, Ryder SD, James MW, et al. Obesity and type 2 diabetes are important risk factors underlying previously undiagnosed cirrhosis in general practice: a cross-sectional study using transient elastography. Aliment Pharmacol Ther 2018;47:504-15. [Crossref] [PubMed]
- Harris R, Card TR, Delahooke T, et al. Obesity Is the Most Common Risk Factor for Chronic Liver Disease: Results From a Risk Stratification Pathway Using Transient Elastography. Am J Gastroenterol 2019;114:1744-52. [Crossref] [PubMed]
- Papatheodoridi M, Hiriart JB, Lupsor-Platon M, et al. Refining the Baveno VI elastography criteria for the definition of compensated advanced chronic liver disease. J Hepatol 2021;74:1109-16. [Crossref] [PubMed]
- Boursier J, Zarski JP, de Ledinghen V, et al. Determination of reliability criteria for liver stiffness evaluation by transient elastography. Hepatology 2013;57:1182-91. [Crossref] [PubMed]
- Papadimitriou P, Garcia-Molina H. Data Leakage Detection. IEEE Transactions on Knowledge and Data Engineering 2011;23:51-63. [Crossref]
- Johnson JM, Khoshgoftaar TM. Survey on deep learning with class imbalance. Journal of Big Data 2019;6:27. [Crossref]
- Zhang S, Li X, Zong M, et al. Efficient kNN Classification With Different Numbers of Nearest Neighbors. IEEE Trans Neural Netw Learn Syst 2018;29:1774-85. [Crossref] [PubMed]
- Reddy KVA, Ambati SR, Reddy YSR, et al. AdaBoost for Parkinson's Disease Detection using Robust Scaler and SFS from Acoustic Features. 2021 Smart Technologies, Communication and Robotics (STCR); 09-10 October 2021; Sathyamangalam, India. IEEE; 2021:1-6.
- Gupta S, Namdev U, Gupta V, et al. Data-driven Preprocessing Techniques for Early Diagnosis of Diabetes, Heart and Liver Diseases. 2021 Fourth International Conference on Electrical, Computer and Communication Technologies (ICECCT); 15-17 September 2021; Erode, India. IEEE; 2021:1-8.
- International classification of diseases for mortality and morbidity statistics (11th Revision): World Health Organization; Available online: https://icd.who.int/browse11/l-m/en.
- Hickman PE, Koerbin G, Potter JM, et al. Choice of Statistical Tools for Outlier Removal Causes Substantial Changes in Analyte Reference Intervals in Healthy Populations. Clin Chem 2020;66:1558-61. [Crossref] [PubMed]
- Cai J, Luo J, Wang S, et al. Feature selection in machine learning: A new perspective. Neurocomputing 2018;300:70-9. [Crossref]
- Bahl A, Hellack B, Balas M, et al. Recursive feature elimination in random forest classification supports nanomaterial grouping. NanoImpact 2019;15:100179. [Crossref]
- Sanai FM, Keeffe EB. Liver biopsy for histological assessment: The case against. Saudi J Gastroenterol 2010;16:124-32. [Crossref] [PubMed]
- West J, Card TR. Reduced mortality rates following elective percutaneous liver biopsies. Gastroenterology 2010;139:1230-7. [Crossref] [PubMed]
- Blanes-Vidal V, Lindvig KP, Thiele M, et al. Artificial intelligence outperforms standard blood-based scores in identifying liver fibrosis patients in primary care. Sci Rep 2022;12:2914. [Crossref] [PubMed]
- Harris R, Card TR, Delahooke T, et al. The XL probe: A luxury or a necessity? Risk stratification in an obese community cohort using transient elastography. United European Gastroenterol J 2018;6:1372-9. [Crossref] [PubMed]
- Docherty M, Regnier SA, Capkun G, et al. Development of a novel machine learning model to predict presence of nonalcoholic steatohepatitis. J Am Med Inform Assoc 2021;28:1235-41. [Crossref] [PubMed]
- Perakakis N, Polyzos SA, Yazdani A, et al. Non-invasive diagnosis of non-alcoholic steatohepatitis and fibrosis with the use of omics and supervised learning: A proof of concept study. Metabolism 2019;101:154005. [Crossref] [PubMed]
- Liu YX, Liu X, Cen C, et al. Comparison and development of advanced machine learning tools to predict nonalcoholic fatty liver disease: An extended study. Hepatobiliary Pancreat Dis Int 2021;20:409-15. [Crossref] [PubMed]
- Graupera I, Thiele M, Serra-Burriel M, et al. Low Accuracy of FIB-4 and NAFLD Fibrosis Scores for Screening for Liver Fibrosis in the Population. Clin Gastroenterol Hepatol 2022;20:2567-2576.e6. [Crossref] [PubMed]
Cite this article as: Bennett L, Mostafa M, Hammersley R, Purssell H, Patel M, Street O, Athwal VS, Hanley KP, Hanley NA, Morling JR, Guha IN; The ID-LIVER Consortium. Using a machine learning model to risk stratify for the presence of significant liver disease in a primary care population. J Med Artif Intell 2023;6:27.