
Efficient glioma grade prediction using learned features extracted from convolutional neural networks
Highlight box
Key findings
• We proposed a convolutional neural network (CNN) architecture that can extract features that are valuable for accurate classification of low-grade and high-grade tumors.
What is known and what is new?
• Previous work has explored the use of complex deep learning models like U-Net, Auto Encoders, and Stacked CNN Architectures for glioma grade prediction.
• The novelty lies in the development of a simple CNN architecture specifically designed to train on individual magnetic resonance imaging sequences. The proposed architecture not only improved performance but also demonstrated faster training and evaluation.
Implication and what should change now?
• It is recommended to consider adjustments in the pre-processing steps to achieve high performance, given the characteristics of the dataset.
Introduction
Brain tumor is an abnormal growth of cells within the brain or the surrounding tissues of the central nervous system (CNS). In the US, there are 23 out of 100,000 population diagnosed with brain tumors in 2011–2015 (1). These tumors can be either benign or malignant (2). Malignant brain tumors require aggressive therapies and are the most challenging to treat. The most common malignant brain tumor in adults is gliomas. Gliomas originate from the glial cells that provide support and protection to neurons and play a crucial role in maintaining the normal functioning of the nervous system. The prevalence of gliomas is approximately 5–10 per 100,000 in the population every year (3). Gliomas are classified into low-grade (least aggressive) and high-grade (most aggressive). The prognosis of a patient with glioma is highly related to the tumor grade (1). Patients with high-grade glioma (HGG) have a poor survival rate, despite treatment options including chemotherapy, radiation therapy, and surgery (4). The survival rate after diagnosis of a brain tumor is 35.8% (5). Especially, patients with the most aggressive brain tumor, glioblastoma multiforme (GBM), have a survival period of 12–16 months, even with advanced treatments (6).
Treatment to a patient with glioma depends on the tumor grade (7,8). Traditionally, glioma grade is determined by pathologists in an invasive manner, by examining a tissue sample under a microscope. This process is expensive, time-consuming, and can have human errors. Therefore, an accurate and robust non-invasive diagnosis method for grade prediction is highly desirable.
In the past, several studies have demonstrated the use of radiomic features to grade gliomas non-invasively from magnetic resonance (MR) images. To classify low-grade glioma (LGG) from HGG, Skogen et al. (9) used histogram-based texture analysis on 95 patients. This study reported a receiver operating characteristic area under the curve (ROC AUC) of 0.910 (9). Tian et al. (10) used a support vector machine (SVM) model to classify grades and reported an accuracy of 98%. In another study, random forest (RF) classifier was used on the radiomic and wavelet-based features and reported an accuracy of 97.54% (11). These studies first extracted the radiomic features from MR images and then these features were used by machine learning (ML) models for determination of glioma grade. It is important to note that ML models, such as SVM, decision trees (DT), RF, and gradient boosting classifier, perform well when the data is limited but require extraction of features. Additionally, these prior studies used hand-engineered features that are straightforward to extract. We believe that the information extracted in these features is limited, thereby, restraining the performance of ML models. Therefore, it is required to build models that learn features directly from MR images.
Recently, several deep learning (DL) models, specifically utilizing convolutional neural networks (CNNs) that learn features from the MR data in a layer-by-layer manner were proposed for glioma grade classification (12,13). For the classification of LGG and HGG, Ertosun and Rubin (14) proposed a CNN and reported an accuracy of 96%. In another study, a CNN and genetic algorithm was proposed by Anaraki et al. (15) and reported an accuracy of 90.9%. In another study, a transfer-learning based approach for glioma grading was proposed by Yang et al. (16), and reported an accuracy of 90%. The CNNs proposed in recent times have the ability to learn features directly from MR images. But it is important to note that these DL methods are data hungry, requiring lots of data to learn robust and meaningful features.
In this paper, we propose a CNN + ML model for accurate grade classification. A CNN model was used to learn features and these learned features are then used by ML models for grade classification. To account for limited data problem, 2D slices were extracted from 3D scans and these 2D slices were used in training CNN model to extract robust features. This approach takes advantage of both CNN model that can learn robust features and ML models that perform well in limited data cases. We compare the proposed approach with the radiomic features and other state-of-the-art CNN models that were used for feature extraction.
Methods
In this section, we first provide the data description and then the different methods utilized for grade classification would be described.
Imaging data description
The dataset used in this work were from the brain tumor segmentation 2018 (BraTS2018) challenge (17,18). BraTS initial focus was only on the evaluation of state-of-the-art methods for the segmentation of brain tumors in multimodal magnetic resonance imaging (MRI) scans (19). Later, the same dataset was being used for other tasks such as grade classification and overall survival rate prediction.
The BraTS 2018 dataset consists of multi-institutional clinically-acquired pre-operative multimodal MRI scans of glioblastoma (GBM/HGG) and LGG (17,18). The dataset consists of 210 patients with HGG and 75 patients with LGG, making it a total of 285 patients. For each patient, the following scans were collected: (I) native (T1); (II) post-contrast T1-weighted (T1Gd); (III) T2-weighted (T2); and (IV) T2 fluid attenuated inversion recovery (FLAIR) volumes. The representative LGG and HGG scans for different sequences were shown in Figure 1. Top row corresponds to HGG scans and bottom row corresponds to LGG scans. Columns 1–4 correspond to FLAIR, T1, T1CE, and T2 scans respectively. The datasets were provided after the following pre-processing steps: (I) co-registered to the same anatomical template; (II) interpolated to the same resolution (1 mm3); (III) skull-stripped; (IV) manual segmentation of tumors by experienced neuro-radiologists (17,18,20).
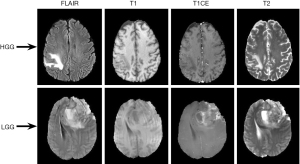
It is important to note that the scans were acquired with different clinical protocols and various scanners from multiple (n=19) institutions. In contrast, the private datasets collected by a single organization vary in the imaging modalities used, the time of data collection, and in the processing techniques employed to clean the data. For these reasons, it is difficult to compare the performance of various algorithms using private datasets.
Radiomic feature extraction
PyRadiomics (21), an open-source package was utilized to extract radiomic features from brain tumor images. The flowchart showing the pipeline of grade classification using radiomic features was shown in Figure 2.
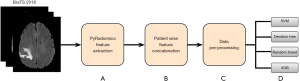
Step A: the initial step involves the extraction of radiomic features, that capture valuable information, from different sequences (T1, FLAIR, T2, T1CE). A total of 110 features were extracted from each sequence, which include First Order Statistics (19 features), Shape-based (3D) (16 features), Gray Level Co-occurrence Matrix (24 features), Gray Level Run Length Matrix (16 features), Gray Level Size Zone Matrix (16 features), Neighbouring Gray Tone Difference Matrix (5 features), and Gray Level Dependence Matrix (14 features) (22).
Step B: subsequently, we have concatenated the features extracted from different sequences corresponding to a single patient, to create a multi-modal feature set. This consolidation enhances the model’s ability to leverage multimodal data for accurate tumor segmentation.
Step C: as part of pre-processing, we stratified split the data into training and testing in the ratio of 75% and 25% respectively. We then employed synthetic minority oversampling technique (SMOTE) oversampling technique (23) on the training data to create a balanced dataset. Subsequently, data normalization has been applied to ensure that the training data is uniformly scaled for faster training of ML models.
Step D: the pre-processed training datasets were given as the input to various ML models, including SVM (24), DT (25), RF (26), and Xtreme gradient boosting (XGBoost) (27) to build the model for glioma classification. Five-fold cross-validation was performed to tune the hyperparameters of ML models. The trained models were then evaluated on the test data.
Learned feature extraction
In this subsection, we detail the methods that were used for extraction of learned features.
Pre-processing
The pre-processing pipeline implemented for extraction of learned features from CNN’s was presented in Figure 3.
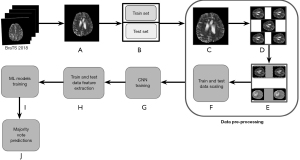
Step A: image extraction
Each patient data consists of four sequences, T1, T1CE, T2, and FLAIR and each sequence is three-dimensional. To account for limited data problem, we first extracted 2D images of size 240×240 from FLAIR, T1CE, and T2 sequences. We excluded T1 sequence due to the limited information present in these images (28). Note that in each 3D sequence, there are 188 slices and only slices from 56 to 136 were included in the study. This range was chosen as it contains the most pertinent information for glioma analysis, avoiding slices that are predominantly black or lacking significant diagnostic features. Note that all the slices from 56 to 136 were included to avoid any data loss, though the neighboring slices may be very similar to each other, leading to redundancy.
Step B: train-test split
We performed patient-level train-test split in the ratio of 75% and 25%. Note that performing patient level split is crucial for avoiding data leakage. Note that the hyperparameters (such as depth of the DT, number of estimators in RF and gradient boosting algorithms, cut-off for defining HGG and LGG) of ML models, were tuned using five-fold cross-validation. Since the dataset is imbalanced, F1-score (harmonic mean of precision and recall) was used as a metric to determine optimal hyperparameters. For instance, to determine optimal cut-off, F1-score was computed at different probability thresholds and the one with the highest F1-score was chosen.
Step C: resizing and cropping
As part of pre-processing, we have resized the extracted images in step A of Figure 3 from 240×240 to 190×190 to reduce the number of trainable parameters. Any further reduction in the size significantly impacted the image quality. We then cropped these resized images to 120×120 to eliminate the external excess black region around the skull part.
Step D: data augmentation
Data Augmentation technique involved generating different versions of the images introducing variations such as rotations, flips. This part of the pre-processing was aimed to enhance model’s ability to generalize well in case of limited data. It is important to note that the data augmentation was performed only on the train set.
Step E: oversampling
To address the problem of class imbalance in the dataset, we have employed oversampling technique. This process involves the replication of minority class data which in our case is LGG, thereby balancing the class distribution and improving the model’s performance.
Step F: data scaling
We have applied normalization on both train and test sets for faster training of CNN models.
Step G: CNN model training
This step involves training different CNN models that were discussed next. The main goal of these CNN models is to learn informative features from the tumor images that are valuable for accurate grade classification.
Step H: CNN feature extraction
Upon CNN model training, we extracted features for both train and test sets. These features provide a rich representation of tumor characteristics.
Step I: training ML models
The extracted features from step H of Figure 3 were used to train various ML models which include the SVM, DT, RF, and XGBoost.
Step J: majority voting
Note that the steps described above from A to I of Figure 3 were performed on 2D images. To determine the grade of a patient, we utilize a majority voting scheme, where the most frequently occurring class of a patient’s feature vectors will be considered as the patient’s grade.
Existing methods
Autoencoder trained on individual sequences for feature extraction
Autoencoder is an unsupervised learning algorithm, mainly used for the task of representation learning (29). The autoencoder architecture used to extract features is presented in Figure 4
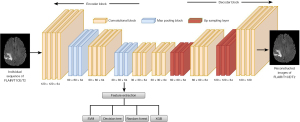
As shown in Figure 4, the network consists of an encoder block and a decoder block. The encoder block comprises of convolutional and max-pooling layers, that are responsible for reducing the input dimensionality, and serve as feature extractor. The decoder block, following the latent space, consists of up-sampling layers to reconstruct the input data from the compressed representation from the encoder block. The autoencoder architecture visually demonstrates the compression and eventual reconstruction of the input image.
Three individual autoencoders were trained, one for each sequence (T1CE, T2, and FLAIR), with the architecture shown in Figure 4. Each individual autoencoder was built with the main goal of reconstructing the input image with minimal error possible, thereby enabling the extraction of meaningful features from the encoder block.
The output of the encoder block was used to extract features from each sequence individually. Once the features were extracted from each sequence, they were concatenated to form a multimodal feature set. These concatenated features were then used to train ML models, SVM, RF, DT, and XGBoost, for glioma grade prediction. Note that the majority voting was utilized to obtain patient-level grade prediction.
CNN trained on stacked sequences for feature extraction
The CNN architecture employed to learn features from stacked T1CE/FLAIR/T2 sequences (30) was shown in Figure 5. The input to the architecture is of shape (120×120×3), where 120×120 represents the dimensions of the image formed after the pre-processing pipeline and 3 represents various sequences (FLAIR, T2, and T1CE) to form a multi-modal input for the 2D-CNN. The architecture comprises a series of convolutional layers, each followed by max-pooling layers to strategically down-sample the spatial dimensions. The flattened output from the convolutional layers leads to two dense layers with rectified linear unit (ReLU) activation function and dropout for regularization. The final output layer consists of a single neuron with a sigmoid activation function for binary classification.
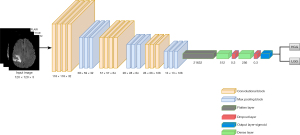
After the 2D-CNN was trained, features were extracted from the dense layer comprising 256 neurons that provide a rich representation of tumor characteristics. These features were then used to train various ML algorithms, SVM, RF, DT, and XGBoost. The majority voting technique was utilized to determine the glioma grade for each patient.
U-Net trained on stacked sequences for feature extraction
The U-Net architecture (31), was first designed and applied in 2015 to process biomedical images. The input to the network consists of stacked sequences of shape (120×120×3), where 120×120 represents the image size and 3 represents different sequences (T1CE, T2, and FLAIR). The U-Net (31) architecture, as shown in Figure 6 consists of encoder-decoder blocks. The encoder block consists of convolutional layers, max-pooling layers for downsampling, and batch normalization to make training faster and more stable reducing issues related to internal co-variant shift (32). On the other hand, the decoder block consists of up-sampling layers to restore the spatial dimensions. The concatenation layer in the decoder blocks played an important role in preserving the spatial features lost during the downsampling process. They combine the feature maps from different levels of the network, ensuring that the final output contains both high-level abstractions and low-level details. The final output layer consists of a single neuron with sigmoid activation function for grade classification.

The U-Net architecture has the advantage of extracting features, that capture the complex patterns present in the multi-modal input data. The features, encapsulating the unique characteristics of tumor regions, were extracted from the dense layer with 256 neurons. These features were then used by ML models and majority voting was used for predicting the grade of a patient.
Proposed method: CNN trained on individual sequences for feature extraction
The proposed CNN architecture trained on individual sequences is presented in Figure 7. The architecture consists of three convolutional layers, each followed by a max pooling layer. The output from convolutional layers was flattened and given as input to dense layer with 512 neurons and ReLU activation function. The output from the dense layer was given to a dropout layer to avoid overfitting issues. While the convolutional layers are specialized in feature extraction and spatial dimension reduction, dense layers are capable of further processing the learned features. The output layer consists of a single neuron with sigmoid activation function for binary classification. This layered structure showcases the gradual transition from feature extraction to classification.

In this proposed method, we pursued a novel approach by training three simple 2D-CNNs, one for each sequence (T1CE/T2/FLAIR). The main objective of these separate CNNs is to capture the sequence-specific patterns in tumor classification resulting in a simplified and focused learning process. Following the training, features learned by each network on individual sequences have been extracted from the dense layer. These features encapsulate the unique characteristics of each sequence, and they were concatenated to form a multi-modal feature set. These concatenated features subsequently were used to train ML models.
Results
The results obtained from different methods described in “Methods” section were presented in Table 1. These results provide valuable insights about the performance of various feature extraction methods. The metrics accuracy (A), precision (P), recall (R), F1-score (F1), specificity (S), and negative predicted value (NPV) were reported for ML models, SVM, DT, RF, and XGBoost. These ML models were trained on features extracted from various techniques, radiomic features, autoencoder trained on individual sequences, CNN trained on stacked sequences, U-Net trained on stacked sequences, and the proposed method which is CNN trained on individual sequences.
Table 1
| Model | SVM (%) | Decision tree (%) | Random forest (%) | XGBoost (%) | |||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A | P | R | F1 | S | NPV | A | P | R | F1 | S | NPV | A | P | R | F1 | S | NPV | A | P | R | F1 | S | NPV | ||||
| Radiomic features | 73 | 68 | 71 | 69 | 80 | 84 | 71 | 63 | 64 | 63 | 85 | 80 | 79 | 74 | 69 | 71 | 85 | 81 | 78 | 72 | 67 | 68 | 85 | 83 | |||
| Individual 2D-AE | 83 | 81 | 71 | 76 | 80 | 84 | 76 | 80 | 71 | 75 | 85 | 92 | 90 | 91 | 90 | 90 | 95 | 90 | 86 | 84 | 83 | 83 | 90 | 88 | |||
| Stacked 2D-CNN | 93 | 92 | 93 | 92 | 90 | 85 | 81 | 80 | 79 | 79 | 90 | 77 | 93 | 92 | 91 | 91 | 95 | 82 | 90 | 90 | 89 | 89 | 90 | 87 | |||
| Stacked U-Net | 98 | 97 | 96 | 97 | 95 | 93 | 98 | 99 | 95 | 97 | 95 | 92 | 98 | 97 | 98 | 98 | 95 | 95 | 99 | 98 | 97 | 98 | 95 | 95 | |||
| Proposed | 98 | 98 | 96 | 97 | 95 | 97 | 89 | 86 | 87 | 86 | 80 | 85 | 98 | 97 | 95 | 96 | 95 | 97 | 98 | 97 | 98 | 98 | 95 | 98 | |||
First row: radiomic features; second row: features learned from an autoencoder network trained on individual sequences; third row: features learned from 2D-CNN model trained on stacked sequences; fourth row: features learned from U-Net model trained on stacked sequences; fifth row: proposed method with CNN trained on individual sequences. Random forest model trained on features learned from autoencoder had an F1-score of 90%, which is 19% higher than radiomic features. The stacked U-Net model had an F1-score of 98%, which is 7% higher than the model trained with features learned by stacked 2D-CNN. The proposed method performed similar to U-Net with F1-score of 98%, but has 14 times less trainable parameters, making it a computationally efficient approach. ML, machine learning; SVM, support vector machine; XGBoost, Xtreme gradient boosting; A, accuracy; P, precision; R, recall; F1, F1 score; S, specificity; NPV, negative predicted value; AE, autoencoder; CNN, convolutional neural network.
The results of ML models trained on radiomic features extracted by PyRadiomics are shown in the first row of Table 1. The best result was obtained by RF algorithm with an F1-score of 71%. Despite our best efforts in tuning hyper-parameters, the results of ML models trained on radiomic features did not improve. This indicates the lack of important information in radiomic features that is required for accurate glioma grade prediction. These results indicate the necessity of learning the features that are relevant for grade classification.
The results of ML models trained on features learned from autoencoder model are shown in the second row of Table 1. The best result was obtained by RF algorithm with a F1-score of 90%, which is about 19% improvement over radiomic feature extraction.
The results of ML models trained on features learned from CNN model trained on stacked sequences are shown in the third row of Table 1. The best result was obtained by SVM with an F1-score of 92%, which is about 2% improvement over autoencoder model.
The results of ML models trained on features learned from U-Net model trained on stacked sequences are shown in the fourth row of Table 1. The best result was obtained by RF and XGBoost models with an F1-score of 98%. This is about 6% improvement over the SVM model trained from features extracted from CNN model trained on stacked sequences. It is important to note that the results obtained from autoencoder method did not yield the best results compared to other CNN models that were trained with a goal of performing classification. It might be because the primary goal of an autoencoder was to reconstruct the image with minimum reconstruction error which might have overshadowed the feature extraction process.
The results of ML models trained on features learned from the proposed model are shown in the fifth row of Table 1. The best result was obtained by XGBoost algorithm with an F1-score of 98%. Furthermore, the proposed method has a specificity of 95% and a negative predictive value (NPV) of 98%. This reflects the model’s reliability in correctly identifying negative cases (LGG). Note that the optimal threshold for the proposed model was 0.464 as determined by five-fold cross-validation. These results indicate that the features extracted from 2D-CNN’s trained on individual sequences contain information that are relevant for accurate grade prediction.
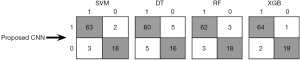
To provide a detailed analysis of the proposed CNN method performance on the test data, confusion matrix for ML models, SVM, DT, RF, XGBoost (XGB) is presented in Figure 8. The label 1 indicates HGG and label 0 indicates LGG. True values are shown in rows and predicted values are shown in columns. The results indicate the ability of the proposed model to accurately classify LGG and HGG. As can be seen in Figure 8, XGBoost, when trained with features extracted from the proposed CNN model has a high number of true positives (TP =64) and a very low number of false negatives (FN =2). This indicates that the proposed model successfully identified majority of positive cases, with only a few instances of misclassification. Such a high TP rate is particularly crucial for accurately detecting the presence of HGG, that is vital for timely and appropriate treatment. The model’s low FN rate further underscores its reliability, minimizing the risk of overlooking patients who require critical medical attention.
To further evaluate the effectiveness of the proposed method, we computed the confidence interval (CI) to quantify the uncertainty of the model’s performance. The F1-score 95% CI of XGBoost algorithm trained on radiomic features is 68±7. The 95% CI of XGBoost algorithm trained on features learned by the autoencoder network is 83±5. The 95% CI of XGBoost algorithm trained on features learned by the stacked 2D-CNN network is 89±4. The 95% CI of XGBoost algorithm trained on features learned by the stacked U-net and the proposed network is 98±1. From these results, it is clear that the 95% CI for the stacked U-net and the proposed network are significantly low compared to the other models. These results highlight the robustness of the features extracted by the proposed network for glioma grade classification.
From the results presented in Table 1, it was clear that the performance of U-Net similar to the proposed method. The comparison of U-Net in terms of number of trainable parameters and training and testing time are given in Table 2.
Table 2
| Method | Train time (sec) | Test time (sec) | Parameters (M) |
|---|---|---|---|
| U-Net | 25,200 | 14.7 | 470 |
| Proposed method | 5,400 | 2.3 | 33 |
The U-Net takes about 7 hours to train and the proposed method takes about an hour and 30 minutes for 20 epochs. The U-Net takes about 14.7 seconds to extract learned features from each test sample, whereas the proposed method takes around 2.3 seconds which is at least 6 times faster compared to U-Net. The U-Net has about 470 million parameters, while the proposed method has 33 million parameters, which is about 14 times lower trainable parameters. sec, seconds; M, millions.
The U-Net utilized had approximately 470 million parameters leading to training time of approximately 7 hours and 14.7 seconds for extraction of learned features from each test sample. On the other hand, the proposed CNN had about 33 million parameters and the training was completed in an hour and 30 minutes and it took around 2.3 seconds to extract learned features from the test set. Note that both the models were run for 20 epochs and all the experiments were conducted on an Intel(R) Xeon(R) Gold 6144 CPU @ 3.50 GHz server with a total RAM of 64 GB. It was evident from Table 2, that the proposed model is simple and has about 14 times less trainable parameters compared to the U-Net. Note that the low parameter count also reduces the risk of overfitting. In addition to its simplicity, the proposed method demonstrated impressive results in determining features that are relevant for accurate glioma grade prediction. In contrast, methods like U-Net, while capable of achieving high performance, come with the drawback of huge number of trainable parameters with a higher computational burden. The proposed method significantly reduced the training and testing time, making it a practical choice for faster model development and evaluation. These observations highlight the trade-off between model complexity, training and testing time, and predictive performance, offering valuable insights for the selection of an appropriate approach.
Discussion
In this study, we proposed a CNN to learn features that are valuable for glioma grade classification. The extracted features are then used to train ML models, SVM, DT, RF, and XGBoost for prediction of grade. The proposed feature extraction network was compared with different architectures including autoencoder network, stacked 2D-CNN, and stacked U-Net.
The model’s effectiveness was assessed based on accuracy, precision, recall, F1-score, specificity, and NPV. The stacked U-Net and the proposed method demonstrated higher performance compared to autoencoder and stacked 2D-CNN networks, highlighting the potential of advanced CNN architectures in effectively predicting glioma grades. This evaluation forms the basis for integrating CNN features with traditional ML models for determining glioma grade.
The performance of using only CNN’s for both feature extraction and classification is presented in Table 3. It can be seen that the performance of proposed CNN network and stacked U-Net are similar and are better compared to autoencoder and stacked 2D-CNN models. However, it is important to note that the performance of the proposed network when used for feature extraction (F1-score =98) is better than using it for grade prediction (F1-score =92). These results demonstrate the significance of using CNN’s for extracting features and then building ML models for grade classification.
Table 3
| Method | A (%) | P (%) | R (%) | F1 (%) |
|---|---|---|---|---|
| Individual 2D-AE | 89 | 87 | 85 | 84 |
| Stacked 2D-CNN | 89 | 89 | 87 | 86 |
| Stacked U-Net | 94 | 93 | 92 | 92 |
| Proposed method | 95 | 94 | 90 | 92 |
The performance of proposed and stacked U-Net models is higher than other CNN networks, but lower compared to using the proposed network for feature extraction and ML model for grade prediction. These results indicate the significance of integrating CNN and ML models. CNN, convolutional neural network; A, accuracy; P, precision; R, recall; F1, F1 score; AE, autoencoder; ML, machine learning.
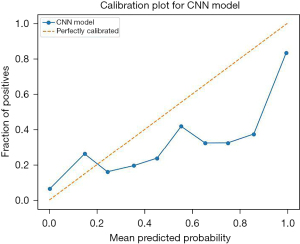
To demonstrate the calibration of the proposed method, calibration plot was presented in Figure 9. It can be observed that the model is well calibrated at lower probabilities and the model has overestimated at higher probabilities (0.7–0.9).
Our findings, as detailed in “Results” section, demonstrate the efficacy of our proposed CNN method in accurately predicting glioma grade. To successfully integrate this technology into clinical practice, a series of steps must be undertaken, including extensive validation of the model against larger and more diverse datasets, ensuring compliance with regulatory standards, and training healthcare professionals to effectively utilize this technology. Furthermore, the integration of our model into existing diagnostic workflow would necessitate collaboration with medical practitioners to ensure seamless implementation.
Conclusions
In this work, we explored various feature extraction methods for glioma grade classification on the BraTS2018 dataset. The feature extraction techniques include radiomic features, autoencoder trained on individual sequences, CNN trained on stacked sequences, U-Net trained on stacked sequences, and CNN trained on individual sequences (proposed method). The ML methods include SVM, DT, RF, and XGBoost that were trained on radiomic and learned features. The performance of ML models trained with learnable features achieved at least a 19% higher F1-score compared to the models trained with radiomic features. The ML models trained with features extracted from the U-Net model had achieved an F1-score of 98%, which is at least 6% higher compared to the models trained with features extracted from autoencoder and stacked CNN network. The proposed method performed similar to U-Net, but with about 14 times less trainable parameters and 6 times faster in extracting features, making it a simple and a computationally efficient approach. In conclusion, autoencoder model focuses on the image reconstruction task and was not an efficient approach for learning features valuable for glioma grading. In addition, features learned from the model trained on individual sequences performed better than the features learned from models trained on stacked sequences. These insights pave a way for tailored treatment strategies and the ultimate goal of improving patient care.
Acknowledgments
Funding: None.
Footnote
Peer Review File: Available at https://jmai.amegroups.com/article/view/10.21037/jmai-23-161/prf
Conflicts of Interest: Both authors have completed the ICMJE uniform disclosure form (available at https://jmai.amegroups.com/article/view/10.21037/jmai-23-161/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. Institutional Review Board (IRB) approval and informed consent are not suitable for this article as it does not involve any human experiment.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Ostrom QT, Gittleman H, Truitt G, et al. CBTRUS Statistical Report: Primary Brain and Other Central Nervous System Tumors Diagnosed in the United States in 2011-2015. Neuro Oncol 2018;20:iv1-iv86. [Crossref] [PubMed]
- Lapointe S, Perry A, Butowski NA. Primary brain tumours in adults. Lancet 2018;392:432-46. [Crossref] [PubMed]
- Wen PY, Kesari S. Malignant gliomas in adults. N Engl J Med 2008;359:492-507. [Crossref] [PubMed]
- Behin A, Hoang-Xuan K, Carpentier AF, et al. Primary brain tumours in adults. Lancet 2003;361:323-31. [Crossref] [PubMed]
- Ostrom QT, Cioffi G, Gittleman H, et al. CBTRUS Statistical Report: Primary Brain and Other Central Nervous System Tumors Diagnosed in the United States in 2012-2016. Neuro Oncol 2019;21:v1-v100. [Crossref] [PubMed]
- Chen J, McKay RM, Parada LF. Malignant glioma: lessons from genomics, mouse models, and stem cells. Cell 2012;149:36-47. [Crossref] [PubMed]
- Cairncross G, Wang M, Shaw E, et al. Phase III trial of chemoradiotherapy for anaplastic oligodendroglioma: long-term results of RTOG 9402. J Clin Oncol 2013;31:337-43. [Crossref] [PubMed]
- Weller M, van den Bent M, Tonn JC, et al. European Association for Neuro-Oncology (EANO) guideline on the diagnosis and treatment of adult astrocytic and oligodendroglial gliomas. Lancet Oncol 2017;18:e315-29. [Crossref] [PubMed]
- Skogen K, Schulz A, Dormagen JB, et al. Diagnostic performance of texture analysis on MRI in grading cerebral gliomas. Eur J Radiol 2016;85:824-9. [Crossref] [PubMed]
- Tian Q, Yan LF, Zhang X, et al. Radiomics strategy for glioma grading using texture features from multiparametric MRI. J Magn Reson Imaging 2018;48:1518-28. [Crossref] [PubMed]
- Kumar R, Gupta A, Arora HS, et al. CGHF: A Computational Decision Support System for Glioma Classification Using Hybrid Features (radiomics based stationary wavelet) over BraTS 2018 Dataset. IEEE Access 2020;8:79440-58.
- LeCun Y, Bottou L, Bengio Y, et al. Gradient-based learning applied to document recognition. Proceedings of the IEEE 1998;86:2278-324. [Crossref]
- LeCun Y, Bengio Y, Hinton G. Deep learning. Nature 2015;521:436-44. [Crossref] [PubMed]
- Ertosun MG, Rubin DL. Automated Grading of Gliomas using Deep Learning in Digital Pathology Images: A modular approach with ensemble of convolutional neural networks. AMIA Annu Symp Proc 2015;2015:1899-908. [PubMed]
- Anaraki AK, Ayati M, Kazemi F. Magnetic resonance imaging-based brain tumor grades classification and grading via convolutional neural networks and genetic algorithms. Biocybern Biomed Eng 2019;39:63-74. [Crossref]
- Yang Y, Yan LF, Zhang X, et al. Glioma Grading on Conventional MR Images: A Deep Learning Study With Transfer Learning. Front Neurosci 2018;12:804. [Crossref] [PubMed]
- Weninger L, Rippel O, Koppers S, et al. Segmentation of Brain Tumors and Patient Survival Prediction: Methods for the BraTS 2018 Challenge. In: Crimi A, Bakas S, Kuijf H, et al. editors. Brainlesion: Glioma, Multiple Sclerosis, Stroke and Traumatic Brain Injuries. BrainLes 2018. Lecture Notes in Computer Science, Springer, Cham; 2018;11384.
- Bakas S, Reyes M, Jakab A, et al. Identifying the best machine learning algorithms for brain tumor segmentation, progression assessment, and overall survival prediction in the brats challenge. arXiv preprint 2018. arXiv:1811.02629.
- Menze BH, Jakab A, Bauer S, et al. The Multimodal Brain Tumor Image Segmentation Benchmark (BRATS). IEEE Trans Med Imaging 2015;34:1993-2024. [Crossref] [PubMed]
- Bakas S, Akbari H, Sotiras A, et al. Advancing The Cancer Genome Atlas glioma MRI collections with expert segmentation labels and radiomic features. Sci Data 2017;4:170117. [Crossref] [PubMed]
- van Griethuysen JJM, Fedorov A, Parmar C, et al. Computational Radiomics System to Decode the Radiographic Phenotype. Cancer Res 2017;77:e104-7. [Crossref] [PubMed]
- Lee T, Song K, Sohn B, et al. A Radiomics-Based Model with the Potential to Differentiate Growth Hormone Deficiency and Idiopathic Short Stature on Sella MRI. Yonsei Med J 2022;63:856-63. [Crossref] [PubMed]
- Fernández A, Garcia S, Herrera F, et al. Smote for learning from imbalanced data: progress and challenges, marking the 15-year anniversary. Journal of Artificial Intelligence Research 2018;61:863-905. [Crossref]
- Noble WS. What is a support vector machine? Nat Biotechnol 2006;24:1565-7. [Crossref] [PubMed]
- Song YY, Lu Y. Decision tree methods: applications for classification and prediction. Shanghai Arch Psychiatry 2015;27:130-5. [PubMed]
- Biau G, Scornet E. A random forest guided tour. TEST 2016;25:197-227. [Crossref]
- Chen T, He T, Benesty M, et al. Xgboost: extreme gradient boosting. R package version 0.4-2, 2015:1-4.
- Maccioni F, Bruni A, Viscido A, et al. MR imaging in patients with Crohn disease: value of T2- versus T1-weighted gadolinium-enhanced MR sequences with use of an oral superparamagnetic contrast agent. Radiology 2006;238:517-30. [Crossref] [PubMed]
- Xu W, Keshmiri S, Wang G. Adversarially approximated autoencoder for image generation and manipulation. IEEE Transactions on Multimedia 2019;21:2387-96. [Crossref]
- Albawi S, Mohammed TA, Al-Zawi S. Understanding of a convolutional neural network. 2017 International Conference on Engineering and Technology (ICET), IEEE; 2017:1-6.
- Jing J, Wang Z, Rätsch M, et al. Mobile-Unet: An efficient convolutional neural network for fabric defect detection. Textile Research Journal 2022;92:30-42. [Crossref]
- Jung W, Jung D, Kim B, et al. Restructuring batch normalization to accelerate CNN training. Proceedings of Machine Learning and Systems 2019;1:14-26.
Cite this article as: Yathirajam SS, Gutta S. Efficient glioma grade prediction using learned features extracted from convolutional neural networks. J Med Artif Intell 2024;7:2.


