
Machine learning methods application: generating clinically meaningful insights from healthcare survey data
Highlight box
Key findings
• A machine learning (ML)-based pipeline with natural language processing (NLP), supervised machine learning (SML), and statistical analysis can be used to generate meaningful insights from healthcare survey data.
What is known and what is new?
• By comparing survey language from the benchmark source and the target source, NLP was applied here to draw insights on respondent’s healthcare experience in the framework of USA quality of care.
• This work ensembled various ML models to predict a patient’s sentiment of their healthcare experience. Classification and feature importance calculations were used to identify the key features that have the most predictive power. Post-process statistical methods were used to generate population level patient tendencies.
What is the implication, and what should change now?
• This work showcased novel application of NLP to healthcare surveys with broad topics to generate the focal topic of interest based on the benchmark survey language of choice.
• The method outlined here demonstrated macroscopic insights from survey data and the leading features that determine survey respondent experiences can be derived with a pipeline combining NLP with SML and introduced a novel method in how healthcare survey data may be digested.
Introduction
With recent accelerations in general computing power (1) and large reductions in the cost of computation, machine learning (ML) has become an integral part of many industries. One benefit of ML applications at scale is the creation of personalized models that can deliver insights at the individual level and improve the overall patient experience. This coincides with a growing demand for personalized, holistic patient centered healthcare. There is a large international demand for personalized care amongst patients suffering from cardiovascular disease, to narrow the gap between the patient experience and a healthcare provider’s own views on the quality of care it provides (2). Previously, it has been shown that hospitals can significantly improve the patient experience by understanding the individual patient journey (3). Application of contemporary technologies like ML for personalized care could not only enhance a patient’s experience but also improve a provider’s abilities by providing tools that can help physicians make decisions using patient level inputs on ML models trained on whole populations (4).
The healthcare industry has often used surveys to understand patient experiences, aiming to improve overall healthcare quality (5-7). Natural language processing (NLP) techniques have been used to make survey analysis more efficient such as by processing patient self-written comments (8). Supervised machine learning (SML) has been used to classify patients’ comments as positive or negative. This work aims to further bridge these separate fields, healthcare surveys and ML, to help capture critical insights and that can be used to provide more personalized patient care.
This analysis applies data science methods to the Beyond Intervention survey (Abbott Laboratories, Santa Clara, CA, USA), which sought to characterize the patient experience across coronary artery disease (CAD) and peripheral artery disease (PAD) through its implementation of three surveys (9-11). The Beyond Intervention survey used in this work explores the perceptions and experiences of 1,289 patients suffering from CAD or PAD, 408 healthcare providers, and 173 healthcare leaders across thirteen countries (10). Key findings from this survey highlight that CAD and PAD patients are more likely to rate their healthcare experience more negatively than their providers and respective healthcare administrative leaders would. By using the Beyond Intervention survey and data-driven insights, and leveraging recent advances in ML, patients’ perceptions of quality of care for CAD and PAD can be elucidated. This can in turn help the healthcare industry develop personalized care that has the potential to improve patient experiences.
Methods
A three-stage framework was proposed in this work analyzing patient survey data to gain insights on indicating factors of patient-perceived quality of care. Section “NLP” describes the first stage, whereby the most relevant Beyond Intervention survey questions relating to patient quality of care were extracted (12). Section “Classification” describes the second stage, during which patient characteristics and feature sets drive the responses to questions extracted in section “NLP” by building SML models that can predict patient responses. Finally, section “Statistical analysis using ML outputs” shows how population likelihoods per values of the patient features can be calculated.
NLP
To algorithmically identify specific Beyond Intervention survey questions upon which to build SML models, NLP techniques were implemented. In the domain of NLP, transformer-based models are considered state-of-the-art and quite popular due to their impressive performance and breath of applications. There are two families of transformer-based models: bidirectional encoder representations from transformers (BERT) and generative pretrained transformer (GPT). These model families focus on distinctive aspects of the transformer architecture: encoder and decoder (13). Given the goal of this work to identify the most relevant survey questions based on a standard benchmark, BERT-based model, specifically, RoBERTa that emphasizes on the encoder perspective of transformer language models was used.
The Hospital Consumer Assessment of Healthcare Providers and Systems (HCAHPS) survey was used as a benchmark to identify Beyond Intervention survey questions relevant to U.S. quality-of-care metrics as described below. The HCAHPS survey is a national, standardized patient satisfaction survey instrument developed by the Centers for Medicare & Medicaid Services (CMS) to quantify patients’ perspectives of their hospital experience. HCAHPS provides information on patients’ experiences in the hospital facility and environment, including care received from nurses and doctors and post-discharge details. From the over 300 questions included in the Beyond Intervention survey, HCAHPS was referenced to extract specific Beyond Interventions questions that were relevant to quality-of-care metrics in the U.S. healthcare system.
NLP data preparation
Data preparation was implemented as the first step in the NLP model to extract related information and eliminate irrelevant data to ensure the efficiency and performance of the system. The relevant language corpus was obtained from the CMS HCAHPS survey and Beyond Intervention survey by eliminating survey questions not explicitly asking about patient experience. Specifically, from the raw survey verbiage, questions related to patient demographics, socioeconomics, disease progression, comorbidities, and symptom information were removed. Standard NLP techniques used in the data preprocessing phase, such as stemming, lemmatization, de-capitalization and removing stop words and low-frequency words were considered but ultimately not needed, due the use of a state-of-the-art bidirectional transformer-based BERT language model. Transformer based models, such as BERT, have been trained on raw text and do not need such preprocessing techniques. Feeding raw text to the transformer-based models preserves the most semantics from the language and produces better results in language modeling tasks.
Language modeling
After the target language corpus was identified for each survey, NLP techniques were applied to extract information from survey questions. Different types of NLP language models were evaluated to create embedding space of the survey languages. Several top-performing transformer-based language models were compared, including the original BERT, XLNet, and RoBERTa models. Since the objective of the current analysis was to identify similar languages from two corpora, after evaluating the performance and the original language model training goals, a pre-trained RoBERTa model was adopted (14). Compared to the original BERT model, RoBERTa eliminated one of the original BERT model’s training objectives, next sentence prediction training, which not only optimized the model performance but also fit the objective of this work better. RoBERTa model can retain the semantics of the raw verbiage better and hence allows for more precise representation of the two sets of survey questions.
The RoBERTa model was used to create a contextualized accurate representation of the survey questions. The words in each question were represented by tokens, including information such as sub-words, which are parts of larger words that indicate context in the word. For example, “ing” in “responding” implies that “a response” is occurring. The tokens were then mapped to embeddings where semantic meanings of the individual words and sub-words were captured. Position embeddings were then added to the token embeddings to incorporate the information about the position of each token within the sequence. These embeddings enabled the model to understand the order of words and their relationships in the sentence. The final representation for each token was obtained by summing the corresponding token and position embeddings. This combined representation was then fed into the Transformer layers, which model the contextual relationships between the tokens. Those steps generate survey language embeddings for both language corpora, as shown in Figure 1.

Compare proximity of embedding space
After data preprocessing and language modeling, vector representations were created for each survey question in the Beyond Intervention (target) and HCAHPS (reference) survey. The cosine similarity score was calculated on each target-reference survey question pair to quantitatively estimate the similarity between the survey language used between the two surveys. The two question pairs with the highest cosine similarity scores are selected for analysis, and are labeled as Q1 and Q2. Q1 and Q2 both have binary response options of “Yes” or “No”.
Classification
Having identified two Beyond Intervention survey questions of interest in section “Compare proximity of embedding space”, this work used these two questions as target variables for two SML pipelines, thereby casting this analysis into a binary classification problem. The goal in this step was to build an accurate classification model that can predict a sample patient’s response to a given question of interest, and then run a feature importance analysis to identify the key patient features that drive patients’ responses and the degree to which they matter.
Classification data preparation
To build a proper classifier, the Beyond Intervention survey data was processed into the appropriate format. The Beyond Intervention survey dataset consisted of 331 columns separated into 69 patient characteristics and 262 patient responses. Of the 69 patient characteristics, 40 were dropped, as they were redundant and/or highly correlated with other variables, mostly single valued and irrelevant, or contained over 50% missing data. The resulting 29 variable patient characteristic dataset contained ten demographic/socioeconomic, one geographic, and eighteen preexisting health status variables. To account for differences in patient experience that are due to different geographies and healthcare systems, the Beyond Intervention dataset was split based on country of the survey respondent. Three countries were selected to proceed with building three different input datasets and three different SML classifiers. The countries selected were the United States of America (USA) (N=150), United Kingdom (N=120), and India (N=125) based on data completeness and utilization of diverse geographical representation.
The input datasets have categorical values. Therefore, label encoding was used to convert the input feature values to integer representations. When the order of values was relevant, such as age brackets, order was preserved. Label encoding was used, as opposed to one hot encoding, to reduce model complexity by reducing the number of input variables used. However, eighteen out of our 28 features had binary values, and were effectively one hot encoded. Moreover, target variables were observed to have an imbalance in the patient response distribution for Q1 in all three countries, with more patients responding “No” in each case. Up-sampling with Synthetic Minority Oversampling Technique (SMOTE) was used for Q1 to create virtual samples in the minority class in the training datasets via linear interpolation of the minority class samples to balance the number of “Yes” and “No” responses (15).
Figure 2 shows the histograms of the USA input dataset for each of the 28 input variables separated by class. The figure shows that there is little one-dimensional separation between the two classes across many of the input features. The USA input dataset is also summarized in tabular form in Table 1. However, by using SML classifiers, this work looked for class separation in 28-dimensional feature space and then calculated feature importance. This was preferred over calculating feature importance solely from the input dataset by measuring the degree of class separation for different values of each feature, only considering separation in the one-d feature space of each feature individually.
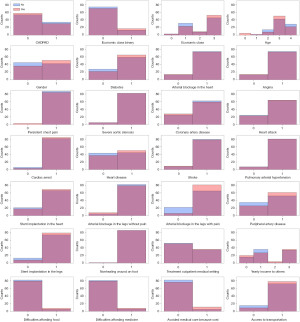
Table 1
| Characteristics | Overall (N=157) |
|---|---|
| Age, n (%) | |
| 35–44 years | 43 (27.4) |
| 45–54 years | 24 (15.3) |
| 55–64 years | 27 (17.2) |
| 65–74 years | 43 (27.4) |
| 75+ years | 20 (12.7) |
| Gender, n (%) | |
| Female | 68 (43.3) |
| Male | 89 (56.7) |
| Diabetes, n (%) | |
| No | 77 (49.0) |
| Yes | 80 (51.0) |
| Hypertension, n (%) | |
| No | 62 (39.5) |
| Yes | 95 (60.5) |
| Hypercholesterolemia, n (%) | |
| No | 61 (38.9) |
| Yes | 96 (61.1) |
| Prior CAD diagnosis, n (%) | |
| No | 56 (35.7) |
| Yes | 101 (64.3) |
| Prior CAD procedure, n (%) | |
| No | 61 (38.9) |
| Yes | 96 (61.1) |
| Recent CAD procedure, n (%) | |
| No | 73 (46.5) |
| Yes | 84 (53.5) |
| Prior PAD diagnosis, n (%) | |
| No | 73 (46.5) |
| Yes | 84 (53.5) |
| Prior PAD procedure, n (%) | |
| No | 84 (53.5) |
| Yes | 73 (46.5) |
| Recent PAD procedure, n (%) | |
| No | 110 (70.1) |
| Yes | 47 (29.9) |
| Prior CAD-related diagnosis, n (%) | |
| No | 18 (11.5) |
| Yes | 139 (88.5) |
| Prior CAD-related medical event, n (%) | |
| No | 60 (38.2) |
| Yes | 97 (61.8) |
| Prior CAD-related symptoms, n (%) | |
| No | 58 (36.9) |
| Yes | 99 (63.1) |
| Prior PAD-related diagnosis, n (%) | |
| No | 30 (19.1) |
| Yes | 127 (80.9) |
| Prior PAD-related medical event, n (%) | |
| No | 100 (63.7) |
| Yes | 57 (36.3) |
| Prior PAD-related symptoms, n (%) | |
| No | 43 (27.4) |
| Yes | 114 (72.6) |
| Comfort with health-tech, n (%) | |
| Comfortable | 32 (20.4) |
| Somewhat comfortable | 49 (31.2) |
| Somewhat uncomfortable | 24 (15.3) |
| Uncomfortable | 12 (7.6) |
| Very comfortable | 27 (17.2) |
| Very uncomfortable | 13 (8.3) |
| Annual household income, n (%) | |
| $25,000–$99,999 | 79 (50.3) |
| <$25,000 | 28 (17.8) |
| $150,000+ | 19 (12.1) |
| $100,000–$149,999 | 31 (19.7) |
| Education level, n (%) | |
| Associate degree | 20 (12.7) |
| Bachelor’s degree | 44 (28.0) |
| Doctorate | 4 (2.5) |
| High school diploma | 21 (13.4) |
| Master’s degree | 34 (21.7) |
| Some college coursework completed | 24 (15.3) |
| Some grade school | 2 (1.3) |
| Technical or occupational certificate | 8 (5.1) |
| Local population density, n (%) | |
| Rural | 44 (28.0) |
| Suburban | 58 (36.9) |
| Urban | 55 (35.0) |
| Insurance type, n (%) | |
| No insurance | 4 (2.5) |
| Private insurance | 79 (50.3) |
| Public insurance | 74 (47.1) |
| Race, n (%) | |
| African American | 15 (9.6) |
| Asian | 7 (4.5) |
| Hispanic | 15 (9.6) |
| Native American or Pacific Islander | 2 (1.3) |
| Other | 8 (5.1) |
| White | 110 (70.1) |
| Experience with telemedicine, n (%) | |
| Very high | 38 (24.2) |
| High | 37 (23.6) |
| Moderate | 21 (13.4) |
| Low | 53 (33.8) |
| None | 8 (5.1) |
| Experience with wearable monitors, n (%) | |
| Very high | 21 (13.4) |
| High | 8 (5.1) |
| Moderate | 46 (29.3) |
| Low | 67 (42.7) |
| None | 15 (9.6) |
| Experience with implantable monitors, n (%) | |
| Very high | 11 (7.0) |
| High | 12 (7.6) |
| Moderate | 14 (8.9) |
| Low | 64 (40.8) |
| None | 56 (35.7) |
| Experience with healthcare mobile apps, n (%) | |
| Very high | 15 (9.6) |
| High | 18 (11.5) |
| Moderate | 35 (22.3) |
| Low | 50 (31.8) |
| None | 39 (24.8) |
| Experience with medical alert devices, n (%) | |
| Very high | 14 (8.9) |
| High | 16 (10.2) |
| Moderate | 8 (5.1) |
| Low | 113 (72.0) |
| None | 6 (3.8) |
| Experience with remote monitoring devices, n (%) | |
| Very high | 21 (13.4) |
| High | 29 (18.5) |
| Moderate | 30 (19.1) |
| Low | 53 (33.8) |
| None | 24 (15.3) |
| Financial hardships, n (%) | |
| Food/water/medicine/healthcare | 36 (22.9) |
| None | 96 (61.1) |
| Other | 14 (8.9) |
| Transportation | 11 (7.0) |
| Type of doctor for CAD/PAD, n (%) | |
| Cardiologist | 88 (56.1) |
| Cardiologist & vascular | 17 (10.8) |
| General | 18 (11.5) |
| Other | 24 (15.3) |
| Vascular | 10 (6.4) |
CAD, coronary artery disease; PAD, peripheral artery disease.
Model training and validation
To select which SML classifier algorithm to use to build a patient survey response model, four different algorithms were tested on Q1 using the USA dataset. These included Sci-Kit Learn’s implementations of Gaussian Process (16), Support Vector Machine (17) using a radial basis function (RBF), Random Forest (18), and Gradient Boosted Classifier (19). Model training was performed by optimizing hyperparameter values by scaling hyperparameter space for each classifier using Sci-Kit Learn’s GridSearchCV module with five-fold cross validation. Mean accuracies, F1-scores, and receiver operating characteristic (ROC)-area under the curve (AUC) for the test and training splits were then obtained.
High mean classification metrics for all models over the training splits were found. This indicates that the models may be overfitting. However, careful overfitting analysis was carried out by scanning hyperparameter space for each model and identifying the parameter values where test accuracy no longer increases as train accuracy increases, as exemplified in Figure 3. Figure 3 shows train and test accuracies for the Gradient Boosting classifier as a function of the hyperparameter “max_leaf_nodes”. The test accuracy increases until “max_leaf_nodes” equals sixteen, at which point the train accuracy is well in the high 90s. The test splits have metrics in the mid 60s to the low 70s. The performance metrics are summarized in Table 2. By comparing these metrics for the different classifiers, a clear algorithm that outperforms the others was not identified, as the metrics were comparable across algorithms. Hence, a stacked classifier, a combination of each classifier’s outputs as inputs into a logistic regression model, was included. Figure 4 shows the model architecture for the final stacked classifier. Although the ensemble stacking does not increase classification metrics, it becomes an important part of the feature importance calculation, which will be demonstrated in the following section.

Table 2
| Classifier | Mean accuracy | Mean F1 | Mean ROC-AUC |
|---|---|---|---|
| Test set | |||
| Support vector machines (RBF) | 0.66 | 0.64 | 0.72 |
| Gaussian process | 0.70 | 0.66 | 0.56 |
| Random forest | 0.63 | 0.63 | 0.65 |
| Gradient boosting | 0.66 | 0.66 | 0.67 |
| Stacked | 0.65 | 0.66 | 0.72 |
| Train set | |||
| Support vector machines (RBF) | 0.98 | 0.98 | 0.99 |
| Gaussian process | 0.99 | 0.99 | 0.99 |
| Random forest | 0.91 | 0.91 | 0.97 |
| Gradient boosting | 0.99 | 0.99 | 0.99 |
| Stacked | 0.98 | 0.98 | 0.99 |
F1 is the harmonic mean of the precision and the recall, where precision = (number of true positives) / (number of true positive + number of false positives) and recall = (number of true positives) / (number of true positives + number of false negatives). ROC-AUC, area under the receiver operating characteristic curve; RBF, radial basis function.

Feature importance
Having built five predictive models which can approximate a given patient’s response to the Beyond Intervention survey question of interest, using the Shapley Additive ExPlanations (SHAP) (20) library, five explainer models were constructed to quantify input feature impact on model prediction output. The resulting mean SHAP values, or quantifications of mean feature impact on model prediction, were calculated and are depicted in Figure 5. The figure illustrates that the four individual models show two general groupings in selecting which input features are the most impactful. Specifically, the figure shows that the Gaussian Process and Support Vector Machine explainer models tend to agree on feature importance, as they show the same top nine features, with some small differences in feature order. Similarly, the Gradient Boosting and Random Forest explainer models show agreement on their top two most impactful features. These two groups can be labeled as kernel-based models (Gaussian Process and Support Vector Machine), and tree-based models (Gradient Boosting and Random Forest). These two groups also agree with each other, in that the top three features of the kernel-based models, “Age”, “Economic class”, and “Yearly income to others”, show up in the top six or top seven features for Gradient Boosting and Random Forest, respectively. Figure 5 shows that the most impactful feature from both tree-based classifiers had a minor impact for the kernel-based models.

Figure 5 also shows that the stacked model combines the most impactful features of its four individual input models, effectively averaging over their respective SHAP values. The stacked model shows five leading impactful features after which there is a large change in the gradient of the feature impact curve. By the sixth feature, the feature impact has decreased more than 2 times that of the leading feature. Ensemble model stacking for SML classifiers is a robust method of ensuring that important information is not lost, when performing feature importance calculations, due to differences in decision boundary resolution between models. While SHAP values from the stacked model suggest a more comprehensive calculation, the Gaussian Process model was subsequently used for simplicity in calculating likelihood of patient response in section “Statistical analysis using ML outputs”.
Statistical analysis using ML outputs
The likelihood of patient response per value of each leading impactful feature was quantified by,
Where is the likelihood that a randomly sampled patient that has the jth value of the ith feature will respond “No” to the key question of interest, and is the ratio of “No” to “Yes” density distribution values for jth value of the ith feature. However, to ensure that the measurements are statistically significant, two criteria were created:
- Density values plus or minus the standard error must not overlap.
- Density values plus or minus the standard error must be above a minimum floor threshold.
The minimum floor, per feature value, was calculated using the “local” standard error, , where is the standard deviation and is the number of samples present in the jth value of the ith feature. This definition is helpful in that the local standard error will go to infinity as goes to zero, and so the minimum floor rises as data quality becomes poor for each feature value separately. Figure 6 illustrates these two criteria being applied to the “Age” feature for the USA input data subset in the Gaussian Process model, separating patients by their Q1 response. The figure shows density distribution values plotted for the two Q1 response groups with error bars indicating the standard error and horizonal bars indicating the minimum floor threshold, or local standard error. Here, we see that we can only calculate meaningful likelihoods from the 65–74 and 75 years and older age brackets. The other three age brackets fail one or both significance criteria.

Results
Based on NLP, the two survey question pairs with the highest cosine similarity scores (0.70 and 0.63) were:
Question pair 1 (Q1):
- HCAHPS: when I left the hospital, I clearly understood the purpose for taking each of my medications.
- Beyond Intervention: my doctor is able to answer all my questions.
Question pair 2 (Q2):
- HCAHPS: during this hospital stay, staff took my preferences and those of my family or caregiver into account in deciding what my health care needs would be when I left.
- Beyond Intervention: my doctor provided me with a personalized treatment plan.
In Q1, the Beyond Intervention response options are binary such that each patient had the choice to select “Yes” or “No” as to whether their doctor was able to answer their questions. Q2 was part of a higher-level question about what would enable patients to have greater trust in doctors. Respondents were asked to rank their top three choices amongst nine possible options, one of which was whether their doctor provided them with a personalized treatment plan. Given that this analysis aimed to understand the factors related to patient satisfaction, the actual ranking of the responses was less important. Hence, the ranking of top three was converted into binary response of ranked (“Yes”) or not ranked (“No”).
The response likelihoods are plotted in Figure 7 for Q1 in the USA data. Here, the likelihood that a randomly-sampled survey respondent will respond “No” vs. “Yes” to Q1 based on various patient features is presented.

Cross model scaling
The modeling process was extended to the United Kingdom and India data subsets for Q1. Using the Gaussian Process model in those cases, we find that the United Kingdom’s Q1 feature explainer model shows the same top three most impactful features as the United States model, and these top three features occur in India’s top five most impactful features. The likelihood analysis was applied for the top three shared most impactful features for all three countries, “Age”, ‘Economic class”, and “Yearly income to others”. The results are presented in Figure 8. Figure 8 shows how patient response likelihood scales across different models, or in this case different countries, by viewing the heat-map column-wise. Amongst United States patients who responded that their “Yearly income to compared to others” was perceived to be below others, most people felt that their doctor was able to answer all of their questions. However, the converse is true for similar patients from the United Kingdom, and patients from India showed no large difference. While these results should be viewed with caution due to limited sample size, the cross-model feature scaling is a powerful method to derive insights from split or stratified datasets.

Extension of method to Q2
The method of extracting survey respondent response likelihoods to a key question by using the patient features that drive the response was extended to Q2, “doctor provides me a personalized treatment plan”. This question is part of a larger survey question where patients were asked to rank their top three factors contributing to trust in one’s doctor among seven options. Table 3 shows the parent questions and the seven response options, including Q2.
Table 3
| Options | Description |
|---|---|
| Option 1 | Doctor provides me with information |
| Option 2 | Doctor spends time with me during appointment |
| Option 3 | Receive frequent follow-up calls |
| Option 4 | Doctor has better tools and technology to diagnose |
| Option 5 | Doctor has higher degree of empathy |
| Option 6 | Improved administration and appointment process |
| Option 7 | Doctor provides me a personalized treatment plan |
Question 2: what would enable you to have greater trust in doctors? Please rank your top 3 where 1 represents the option that would increase your trust in doctors the most, 2 would increase it the second most, etc. Patients were asked to rank their top three factors, out of seven options, in determining trust in their doctor. Question 2 is option seven.
Plotting the distribution of responses for the United States for Q2 shows that there is a large imbalance in counts per response, as shown in Figure 9. This is due to a low number of patients ranking Q2 in their top three factors contributing to doctor trust.
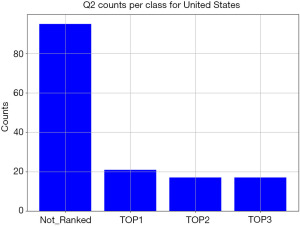
While the method described in section “Classification” is generalizable to a multi-class classification problem, the imbalance in patient responses shown in Figure 9 indicates that the survey data available is not suitable for multi-class classification, as there are not enough samples per class. Hence, Q2 responses were recast into a binary form, where “TOP1”, “TOP2”, and “TOP3” response options were combined into a “ranked” response; if Q2 was not selected within a patient’s top three factors, then it was labelled an “unranked” response.
Using the Gaussian Process algorithm, a binary classifier was trained targeting binary Q2 responses for all three countries. SHAP values were then obtained showing the same top three most impactful input features as those found for Q1. Hence, the same likelihood analysis described for Q1 is applicable to Q2, and a framework similar to Figure 8 can again be used to show the cross-model feature scaling (column-wise) and the intra-model feature scaling (row-wise). Figure S1 shows the cross-model feature scaling for Q2.
Discussion
This analysis proposes a novel method to obtain insights from healthcare survey data using NLP and ML classification, combined with statistical analysis. This method identifies target questions of interest using NLP algorithms which are then used as target outputs for classification models. Different surveys with similar respondent populations can be combined by finding similar survey questions and consolidated using cosine similarity score and language embeddings. Survey sample size could be increased without the requirement of additional surveys. Feature importance calculations and population statistics were then obtained to generate meaningful conclusions.
Extraction of clinical conclusions from the ML pipeline demonstrated in this work would require a final real world validation step where the conclusions are used to inform policy/procedure changes that can lead to better patient quality of care. This would require re-administering of the survey to the respondents. Therefore, this work is a demonstration of the survey analysis method, and the drawing of clinical conclusions are left as future work for the authors.
Feature importance
In this analysis, input feature impact on model prediction has been calculated for four different SML classifiers, all with comparable classification metrics. This showed that classification models with similar accuracies can “disagree” on which input features have the most impact. Moreover, these differences scale with the classifiers’ algorithmic types (e.g., trees or kernels). These differences arise form how each classifier can “carve out” a decision boundary shape in the feature input space. For instance, decision tree splits or kernel function mapping. However, the physical systems being modeled by the classifiers do have one set of driving features that the feature importance calculations are aiming to reveal. Therefore, this work suggests that for classification problems which depend on feature importance/impact calculations, where classification models of different algorithmic types show comparable classification metrics but differ on feature importance calculations, ensemble model stacking is an effective method of averaging over differences in individually calculated decision boundaries, recovering information that may be missed when using a single model.
Emerging methods: generative artificial intelligence (AI)
With the recent developments around large language models (LLMs) and generative AI, there has been a shift in the focus of NLP research from the previous paradigm of training specialized models for specific tasks to adopting large neural networks with many parameters that perform well on a wide variety of tasks. The significant value gain in application of LLMs lies in the fact that they are trained on massive amounts of text data and will be able to capture more subtle nuances and contextual information compared to traditional ML, because they can be used to generate high-quality embeddings and vector representations of input text. In addition, LLMs allow for improved research efficiency with notable ability in text editing and notional tasks. However, the application of emerging LLMs also brings some limitations and challenges (21):
- Hallucinations (factually incorrect or incoherent results) and adversarial jailbreak (LLMs coaxed into overriding safety, privacy guardrails) mitigation. LLMs were trained on enormous dataset to predict next output of the language that makes the most “sense”. However, there is a lack of proven methods to systematically ensure the truthfulness of LLMs’ output and mitigate potential hallucinations and biases. Adversarial jailbreaking of generative language models is also an active field of research. In applications of LLMs in healthcare setting, the importance of maintaining privacy of sensitive care details cannot be overstated.
- Results validation and interpretability. The degree of sentence-matching between the two datasets needs to be numerically quantified. While LLMs can help perform many language tasks, it is important that the results are validated with scientific methods and are meaningful and useful for the application. With additional research in the space, we expect the emergence of novel methods for validation of Generative AI output and methods to interpret why certain results are produced.
- In commercial application of LLMs, especially in healthcare space, there’s additional burdens of ensuring the trustworthiness of AI. Additional factors like usefulness, accountability, fairness, will all need to be evaluated and considered thoroughly (22).
Overall, LLMs and generative AI are areas of ongoing research that hold tremendous promises. However, there are also limitations that come with the significant value. LLMs by no means are best suited for all use cases. The application of traditional NLP methods using patient survey data in this work was more than adequate to accurately capture the semantics of survey language.
Limitations
In the Beyond Intervention survey questions evaluated, there is no distinction made to the specific healthcare provider that delivered care and therefore influenced the patient experience. Specifically, the survey language does not specify a primary care physician versus a specialty physician, nurse, or other healthcare professionals. In contrast, there is a clear distinction made in the HCAHPS survey language. With this limitation in the original survey data, the patients’ experience is taken across the healthcare team as a whole.
Although the Beyond Intervention survey collected patient satisfaction data from across the world, the number of patients surveyed at each individual country was limited. Since the analysis was conducted on country basis, this can lead to model overfitting issues during model training. However, this work does not aim to build a predictive model. The goal here is simply to build an accurate model from which feature importance calculations can be generated to use in statistical comparisons. Moreover, the Beyond Intervention survey was conducted globally in over ten different countries, seeking to measure international patient experience. As different countries may have different indication factors that drive patient quality-of-care, future analyses may be readily conducted by identifying key patient quality-of-care factors from the perspective of other healthcare systems (e.g., the NHS in the UK), using different reference surveys when question-matching via NLP.
Another limitation in our ML pipeline is that it is the correlation of the driving variables and the patient responses that is being calculated, which doesn’t necessarily identify causation. However, all causal relationships are highly correlated but not all correlated ones are causal. Therefore, a high amount of care is required in preparing survey questionnaires so that when the patient characteristic variables are defined, to the best of the survey creators’ clinical foresight, they encompass the causal variables.
Applications
The analysis method developed here can lead to a wide range of applications. Using the NLP portion of this work, researchers can efficiently extract survey questions that align with research interests from the hundreds of questions in the raw survey data. Once a question of interest is selected, the correlation between the survey respondents’ characteristic features and the survey response is quantified, thereby yielding actionable insights. For instance, if a hospital implements a quality-of-care survey and identifies a key survey question as a topic of importance, the method of building a survey respondent classifier and calculating feature importance will inform healthcare officials which patient subgroup(s) are most sensitive to the survey question. In essence, the feature importance step can be viewed as a noise reduction filter, with the noise being the non-sensitive patient characteristics. Healthcare officials can then develop procedure or policy changes to improve the experience of specific patient subgroup(s). At an individual patient level, healthcare providers can further tailor the delivery of care to optimize the patient experience. For example, if a particular patient profile is less likely to feel that their doctor answered their questions or that they were provided an individual treatment plan, a clinician may tailor their discussion with that patient accordingly.
Similar interesting work has been carried out previously in health care, where SML classifiers were constructed to predict insomnia and SHAP was used to identify the leading factors driving the predictions (23). In this work, it is shown that leading SHAP features can be model dependent, and that model stacking helps to reduce this model bias. This in turn helps the real-world application of SML models in the healthcare space, as conclusions drawn from SHAP values can have more robustness.
Conclusions
This work presented a three-step method of extracting useful insights from healthcare survey data leveraging contemporary ML techniques. Firstly, NLP was used to mine the survey and identify questions of interest that align with a prescribed set of questions, here HCAHPS was used as a reference. Then, SML was used to build classifiers that can predict patient question responses, from which explainer models were calculated to find the patient features that drive the patient responses. Lastly, statistical methods were applied to calculate the likelihood of how a randomly sampled patient that falls within a specific feature value of a high impact variable will respond to the survey question of interest. This proposed method can be generalized to different survey question types and different geographical areas of interest with some adaption. The findings from such method will allow healthcare researchers to have a better understanding of the driving factors of patients’ perception to care, and in turn, gain insights on potential development areas of more personalized care.
Acknowledgments
Funding: Abbott funded the
Footnote
Data Sharing Statement: Available at https://jmai.amegroups.com/article/view/10.21037/jmai-24-159/dss
Peer Review File: Available at https://jmai.amegroups.com/article/view/10.21037/jmai-24-159/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://jmai.amegroups.com/article/view/10.21037/jmai-24-159/coif). Q.L., S.T. and J.B.P. are employed by Abbott Laboratories and report salary and stock received from Abbott. D.R. is employed by Microsoft, received consultant for Beyond Intervention Project from 2021–2022 from Abbott, served as Executive Advisory Board member for M42 (2024), Cedars-Sinai Medical Center (2024), NESTcc (2020–2024), AdvaMed (2023–2024), Health Tech Without Borders (2022–2024), Health Wise (2021–2024), Topcon (2023–2024), Mediflix (2023), Blatchford (2023), and holds stock options with Microsoft. The other authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. IRB approval was not applicable as no human subjects were used in this work. Moreover, no personal identifiable data was used.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Dean J. 1.1 the deep learning revolution and its implications for computer architecture and chip design. In2020 IEEE International Solid-State Circuits Conference-(ISSCC) 2020 Feb 16 (pp. 8-14). IEEE.
- West NEJ, Juneja M, Pinilla N, et al. Personalized vascular healthcare: insights from a large international survey. Eur Heart J Suppl 2022;24:H8-H17. [Crossref] [PubMed]
- Gualandi R, Masella C, Viglione D, et al. Exploring the hospital patient journey: What does the patient experience? PLoS One 2019;14:e0224899. [Crossref] [PubMed]
- Johnson KB, Wei WQ, Weeraratne D, et al. Precision Medicine, AI, and the Future of Personalized Health Care. Clin Transl Sci 2021;14:86-93. [Crossref] [PubMed]
- Redding TS, Keefe KR, Stephens AR, et al. Evaluating Factors That Influence Patient Satisfaction in Otolaryngology Clinics. Ann Otol Rhinol Laryngol 2023;132:19-26. [Crossref] [PubMed]
- Pogorzelska K, Chlabicz S. Patient Satisfaction with Telemedicine during the COVID-19 Pandemic-A Systematic Review. Int J Environ Res Public Health 2022;19:6113. [Crossref] [PubMed]
- Satin AM, Shenoy K, Sheha ED, et al. Spine Patient Satisfaction With Telemedicine During the COVID-19 Pandemic: A Cross-Sectional Study. Global Spine J 2022;12:812-9. [Crossref] [PubMed]
- Khanbhai M, Warren L, Symons J, et al. Using natural language processing to understand, facilitate and maintain continuity in patient experience across transitions of care. Int J Med Inform 2022;157:104642. [Crossref] [PubMed]
- Personalized Vascular Care Through Technological Innovation. Abbott Laboratories; 2020. Available online: https://vascular.abbott.com/rs/111-OGY-491/images/BeyondIntervention-WhitePaper.pdf
- Improving patient experience by addressing unmet needs in vascular disease. [cited 2024 Mar 8]. Available online: https://vascular.abbott.com/rs/111-OGY-491/images/BeyondIntervention-WhitePaper-PADsupplement-2021.pdf
- Enhancing Positive Outcomes for Patients. Abbott Laboratories; 2022. Available online: https://vascular.abbott.com/beyond-intervention.html?utm_source=cva-left-cta&utm_medium=website&utm_campaign=beyond-intervention-nov-2021
- Centers for Medicare & Medicaid Services. HCAHPS: Patients’ Perspectives of Care Survey. Cms.gov. 2021. Available online: https://www.cms.gov/Medicare/Quality-Initiatives-Patient-Assessment-Instruments/HospitalQualityInits/HospitalHCAHPS
- Vaswani A, Shazeer N, Parmar N, et al. Attention is all you need. In: Guyon I, Luxburg UV, Bengio S, et al. editors. Advances in Neural Information Processing Systems. Red Hook, NY: Curran Associates, Inc., 2017;30:4765-74.
- Liu Y, Ott M, Goyal N, et al. Roberta: A robustly optimized bert pretraining approach. arXiv preprint arXiv: 1907.11692. 2019 Jul 26.
- Chawla NV, Bowyer KW, Hall LO, et al. SMOTE: synthetic minority over-sampling technique. J Artif Intell Res 2002;16:321-57. [Crossref]
- Rasmussen CE, Williams CKI. Gaussian processes for machine learning. Cambridge, Mass: MIT Press, 2008.
- Cortes C, Vapnik V. Support-vector networks. Mach Learn 1995;20:273-97. [Crossref]
- Ho TK. Random decision forests. Montreal, QC, Canada: Proceedings of 3rd International Conference on Document Analysis and Recognition; 1995:278-82.
- Friedman JH. Greedy function approximation: a gradient boosting machine. Ann Statist 2001;29:1189-232. [Crossref]
- Lundberg SM, Lee SI. A unified approach to interpreting model predictions. In: Guyon I, Luxburg UV, Bengio S, et al. editors. Advances in Neural Information Processing Systems. Red Hook, NY: Curran Associates, Inc., 2017;30:4765-74.
- Cascella M, Montomoli J, Bellini V, et al. Writing the paper “Unveiling artificial intelligence: an insight into ethics and applications in anesthesia” implementing the large language model ChatGPT: a qualitative study. J Med Artif Intell 2023;6:9. [Crossref]
- Blueprint for trustworthy AI implementation guidance and assurance for healthcare coalition for health AI. 2022 [cited 2023 Jul 25]. Available online: https://www.coalitionforhealthai.org/papers/Blueprint%20for%20Trustworthy%20AI.pdf
- Huang AA, Huang SY. Use of machine learning to identify risk factors for insomnia. PLoS One 2023;18:e0282622. [Crossref] [PubMed]
Cite this article as: Lu Q, Taimourzadeh S, Rhew D, Fitzgerald PJ, Harzand A, McCaney J, Prillinger JB. Machine learning methods application: generating clinically meaningful insights from healthcare survey data. J Med Artif Intell 2025;8:4.


