
A comparative study of machine learning algorithms for breast cancer diagnosis
Introduction
Breast cancer is the most prevalent cancer among women worldwide, accounting for 25% of all new cancer diagnoses and 15% of cancer-related deaths (1). Early detection plays a pivotal role in survival rates; the five-year survival rate for early-detected breast cancer exceeds 90%, but it drops drastically to approximately 15% for late-stage detection (2,3). These statistics emphasize the urgent need for enhanced diagnostic and prognostic tools to improve patient outcomes.
Over recent decades, machine learning (ML) techniques have emerged as transformative tools in healthcare, particularly in cancer diagnosis (4). Traditional diagnosis relied heavily on clinicians’ experience, but the ever-growing complexity of medical data—spanning mammographic images, gene expression profiles, and electronic health records—necessitates computational assistance. ML techniques offer an unparalleled ability to process and analyze such high-dimensional datasets, enabling clinicians to differentiate between benign and malignant tumors and predict disease progression (5).
Despite their promise, ML models face numerous challenges in breast cancer diagnosis, such as data complexity, class imbalance, and feature non-linearity. Addressing these challenges is essential for improving clinical reliability and reducing the risks of misdiagnosis (6). This study systematically evaluates the performance of popular ML algorithms, including Support Vector Machines (SVM), Logistic Regression (LR), Random Forest (RF), and Artificial Neural Networks (ANN), for breast cancer diagnosis. Unlike previous reviews, this research focuses on addressing challenges like class imbalance and preprocessing, including tasks like normalizing data, handling missing values, and eliminating redundant features (2,7,8). The goal is to enhance model accuracy and clinical utility. Analyzing non-imaging datasets revealed model accuracy levels ranging from 57% to 92%. The study highlights the importance of preprocessing tasks in improving classification outcomes and underscores the potential of ML techniques like RF and ANN in advancing breast cancer care. By addressing key limitations and emphasizing data quality, this work contributes to the growing body of evidence supporting the use of ML in healthcare, ultimately aiming for more personalized and timely treatments. The findings demonstrate the transformative potential of ML in improving breast cancer diagnosis.
The remainder of this paper is organized as follows: section “Literature review” provides a comprehensive literature review, highlighting prior studies on ML applications for breast cancer diagnosis. Section “Methodology and dataset” describes the datasets, and outlines the experimental methodology, detailing the ML algorithms applied and the evaluation metrics used. Section “Model training and evaluation process” presents the results and comparative analysis of the models. Finally, section “Comparative analysis and discussion” concludes the paper with a discussion of key findings, limitations, and directions for future research.
Literature review
This section provides a contextual analysis of recent studies to situate our comparative research on ML algorithms for breast cancer prediction and diagnosis. The review focuses on both the advancements and the limitations highlighted in prior works, offering a foundation for our experimental analysis. Indeed, ML algorithms have been pivotal in advancing breast cancer diagnosis. They offer enhanced predictive accuracy and can manage complex, large-scale datasets, which are difficult to process using traditional statistical methods. In this perspective, several key ML algorithms have proven effective in breast cancer care:
SVM
SVM is an effective classifier for breast cancer diagnosis, especially in high-dimensional feature spaces. It identifies the optimal hyperplane to separate classes (9) (malignant vs. benign tumors), which is crucial in complex medical datasets with many features. SVM has been particularly useful in distinguishing cancerous and benign cells, as demonstrated in multiple studies. The study of Chen and Jia (10) applies SVM to breast cancer diagnosis, exploring the performance of four commonly used kernel functions across different datasets. Experimental results demonstrate a classification accuracy of 98.25% on the breast cancer dataset. In another work, authors (11) proposed an SVM-based method with feature selection for breast cancer diagnosis, which was tested on the Wisconsin Breast Cancer Dataset (WBCD). The proposed approach achieves a classification accuracy of 99.51% with five features.
Decision trees (DTs)
DTs are known for their interpretability and can handle both numerical and categorical data. Their simplicity and ability to visually represent decision paths make them valuable for clinical environments where transparency is critical for decision-making. In the work of Al-Salihy et al. (12), authors used several DT algorithms, including J48, Function Tree, RF Tree, Alternating Decision Tree (AD Tree), Decision Stump, and Best First, applied to a dataset of 569 cases (357 benign and 212 malignant) with 32 attributes. The algorithms were evaluated using the Waikato Environment for Knowledge Analysis (WEKA) framework. Among these methods, DT models demonstrated strong performance, achieving an average precision of correctly classified cases at 97.7%, highlighting their computational efficiency and diagnostic reliability. Notably, the Decision Stump algorithm yielded the lowest accuracy, at 88.0%, underscoring the variability in effectiveness among different DT classifiers.
RF
RF is an ensemble method that aggregates predictions from multiple DTs to enhance model robustness. Its effectiveness is particularly apparent when handling large and complex datasets, making it useful for breast cancer diagnosis (13). The study of Minnoor et al. (14) highlights RF as a highly effective algorithm for breast cancer diagnosis. Using the WBCD, which comprises 569 cases (212 malignant) and 30 cellular attributes (e.g., radius, texture, concavity), the research aims to develop efficient ML models for tumor classification. RF outperformed other methods, including SVM, DT, Multilayer Perceptron, and K-Nearest Neighbors (KNN). The model was trained on subsets with 16 and 8 features selected via feature selection techniques and further optimized using hyperparameter tuning. The RF algorithm achieved an accuracy of 99.3%, on test datasets.
KNN
KNN classifies tumors based on the proximity of data points in the feature space. This instance-based learning method is simple and effective, especially in smaller datasets (15). The work of Rajaguru (16) investigated the classification of breast tumors using ML techniques, with a focus on comparing the performance of KNN and DT algorithms. Using the Wisconsin Diagnostic Breast Cancer (WDBC) dataset, the research categorizes tumors into benign or malignant. Feature selection is performed using principal component analysis (PCA) to enhance model performance. Standard performance metrics are used to evaluate the algorithms. The results demonstrate that the KNN classifier outperforms the DT algorithm.
Gradient Boosting Machines (GBM)
Gradient Boosting algorithms, including XGBoost, sequentially build models to correct errors made by previous trees. This technique has become a go-to for handling imbalanced datasets, showing notable improvements in diagnostic accuracy. Mangukiya (17) identified XGBoost as a highly accurate algorithm for breast cancer detection but provided limited exploration of other algorithms’ performance or scalability to larger datasets. Similarly, Rabiei (18) reported RF models achieving high sensitivity, while Gradient Boosting models showed superior specificity. However, the reliance on single databases and the absence of genetic data in these studies restricted their generalizability.
Neural Networks (NNs)
NNs excel at recognizing complex, non-linear patterns in data and are widely used in breast cancer prediction. Their ability to process large feature sets and detect intricate patterns often allows them to outperform traditional methods, and in some cases, even human radiologists (19). For example, Gupta (20) explored the use of CNNs for breast cancer prediction, emphasizing edge-based imaging techniques to enhance efficiency in processing time and space. Despite the promising findings, the study lacked detailed training and validation processes, and it failed to address potential limitations in image preprocessing, such as the loss of critical information.
LR
LR is a fundamental algorithm for binary classification, providing a clear and interpretable method to predict probabilities, such as the likelihood that a tumor is malignant or benign (21). The study of Khandezamin et al. (22) presents a two-step method for improving breast cancer diagnosis using fine needle aspiration (FNA) cytology data and ML techniques. In the first step, LR is employed to eliminate less significant features, streamlining the dataset. In the second step, a NN is used to classify tumors as benign or malignant. The method is evaluated on three datasets and the obtained precision reached 96.9%.
Building on the existing body of work, this study aims to address several identified gaps by systematically evaluating the performance of various ML algorithms. Our research goes beyond prior work by utilizing comprehensive metrics, including accuracy, recall, and F1-score, while incorporating robust preprocessing techniques, such as Synthetic Minority Over-sampling Technique (SMOTE), to mitigate the effects of class imbalance. Moreover, we expand on the current focus on imaging data by integrating non-imaging datasets, providing a more holistic approach to breast cancer prediction. Through these measures, we seek to contribute to both the academic and clinical applications of ML in breast cancer care.
Methodology and dataset
Method overview
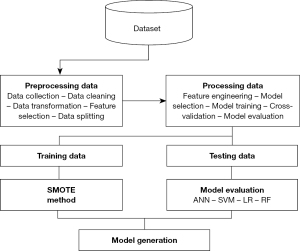
In this work, we compare the performance of four ML algorithms—SVM, RF, LR, and ANN—in breast cancer diagnosis using clinically validated, publicly available tabular dataset with 23 features. Extensive preprocessing was applied to ensure robust model performance, including removing irrelevant or redundant features, imputing missing values using statistical and predictive methods, normalizing continuous features through L2 normalization (23), and addressing class imbalance with the SMOTE (24). Each ML model was trained and evaluated using key performance metrics, such as precision, recall, F1-score, sensitivity, specificity, and area under the curve (AUC). Hyperparameters were fine-tuned to optimize performance, with specific steps including testing linear, radial basis function (RBF), and polynomial kernels for SVM, optimizing tree depth and splitting criteria for RF, performing stepwise variable selection for LR, and adjusting hidden layers and nodes for ANN. The evaluation relied on thresholds derived from the Youden Index (25) to maximize sensitivity and specificity. The experimental workflow, which includes data preprocessing, model training, and evaluation, is outlined in Figure 1, providing a reproducible foundation for this comparative analysis.
Dataset description
Our work relies on a dataset (26) of 716 breast cancer cases diagnosed at the National Institute of Oncology in Rabat, Morocco. This dataset highlights the significant disparities between young women (≤40 years) and older women (>40 years) regarding prognosis and outcomes. The dataset provides valuable information to facilitate breast cancer diagnosis. Key features such as estrogen receptor (ER), progesterone receptor (PR), and tumor size are critical in differentiating benign and malignant cases. These diagnostic features play a pivotal role in training ML models to improve accuracy and reliability in identifying cancer cases. The following table (Table 1) provides an overview of the dataset.
Table 1
| Variables | Description | Type |
|---|---|---|
| ID#patient | Unique identifier for each patient | Numerical |
| Age | Patient’s age category (≤40, >40 years) | Categorical (ordinal) |
| Nulliparity | Whether the patient has never given birth (yes, no) | Categorical (nominal) |
| Oral contraception use | Usage of oral contraceptives (yes, no) | Categorical (nominal) |
| Menopause | Menopausal status of the patient (yes, no) | Categorical (nominal) |
| Familial history of breast cancer | Family history of breast cancer (yes, no) | Categorical (nominal) |
| Number of full-term pregnancies | Number of full-term pregnancies (‘0’, ‘1’, ‘2 to 4’, ‘5 or more’) | Categorical (ordinal) |
| Obesity | Obesity status (yes, no) | Categorical (nominal) |
| Metastatic disease | Presence of metastatic disease (yes, no) | Categorical (nominal) |
| ER | Estrogen receptor status (positive, negative) | Categorical (nominal) |
| PR | Progesterone receptor status (positive, negative) | Categorical (nominal) |
| HER2 | HER2 receptor status (positive, negative) | Categorical (nominal) |
| Tumor size | Tumor size category (T1, T2, T3) | Categorical (ordinal) |
| Lymph nodes | Lymph node involvement (N1, N2, N3, N4) | Categorical (ordinal) |
| Histological type | Type of breast cancer (‘tubular_carcinoma’, ‘invasive_carcinoma_of_no_special_type’, ‘invasive_lobular_carcinoma’, ‘ductal_carcinoma_in_situ’ ‘angiosarcoma’, ‘carcinoma_with_medullary_features’, ‘invasive_mucinous_carcinoma’, ‘metaplastic_carcinoma_of_nst’, ‘carcinoma_with_neuroendocrine_features’, ‘sarcoma’ ‘invasive_papillary_carcinoma’) | Categorical (nominal) |
| Vascular invasion | Presence of vascular invasion (yes, no) | Categorical (nominal) |
| SBR grade | Scarff-Bloom-Richardson grade (SBR I, SBR II, SBR III) | Categorical (ordinal) |
| Surgery type | Type of surgery performed (radical mastectomy, conservative) | Categorical (nominal) |
| Adjuvant chemotherapy | Whether adjuvant chemotherapy was given (yes, no) | Categorical (nominal) |
| Radiotherapy | Whether radiotherapy was administered (yes, no) | Categorical (nominal) |
| Trastuzumab | Whether trastuzumab was administered (yes, no) | Categorical (nominal) |
| Hormone therapy | Whether hormone therapy was administered (yes, no) | Categorical (nominal) |
| progression | Disease progression status (yes, no) | Categorical (nominal) |
Each column may have values like ‘unknown’ or NaN. NaN, not a number.
Research method
To carry out our research effectively, we followed a series of important steps that play a key role in breast cancer diagnosis. The diagram below (Figure 1) outlines these steps.
Data preprocessing
Data preprocessing is an important step in the ML pipeline, significantly affecting model quality and effectiveness. This section covers essential techniques, including data cleaning, handling missing values, data normalization, and addressing imbalanced datasets. Data cleaning analysis focuses on correcting inaccuracies and inconsistencies in the dataset to ensure reliable inputs for modeling. Handling missing values is essential, as incomplete data can skew results; strategies like imputation or removal of missing entries are commonly used (27). Data normalization scales feature to ensure they contribute equally to model performance, especially for algorithms sensitive to feature scales. Additionally, addressing imbalanced data is vital, as class imbalances can lead to biased models. Techniques like SMOTE create synthetic examples of the minority class, balancing the dataset and enhancing model robustness (24). Implementing these preprocessing steps improves data quality, ultimately leading to better performance and accuracy in ML models for breast cancer diagnosis.
Data cleaning analysis
The provided data cleaning process outlines several key steps to prepare a dataset for analysis in the context of breast cancer diagnosis. Here’s an analysis of each step:
- Dropping unlabeled rows: this step ensures that only data points with known outcomes are used, reducing noise and focusing the analysis on relevant, complete data (28). Therefore, we have removed rows in the dataset without labels (i.e., missing outcome or target variable data).
- Dropping the patient ID column: the column labeled ‘id#patient’ was removed from the dataset. This column likely contains unique identifiers for each patient, which do not contribute to the predictive modeling process and can be safely discarded.
- Creating an ‘is_young’ column: a new column named ‘is_young’ is introduced to categorize patients based on age. Women aged 40 years or younger are coded as ‘1’, while those older than 40 are coded as ‘0’. The original ‘age’ column is then dropped, simplifying the dataset and retaining only the binary age classification that may be relevant for further analysis.
- Converting ‘metastatic_disease’ values: this column is transformed, where the value ‘m1’ (indicating the presence of metastatic disease) is converted to ‘1’, and not a number (NaN) (indicating the absence of metastatic disease) is converted to ‘0’. This conversion simplifies the data into a binary form, which is more suitable for ML models.
- Mapping categorical values to numerical values: several categorical variables in the dataset are converted into numerical values using a specific mapping technique. This step is critical for preparing the data for ML algorithms, which require numerical input (29). The values [‘n0’, ‘0’, ‘negative’, ‘no’] are all mapped to ‘0’. This likely represents negative test results or the absence of certain conditions. The values [‘1’, ‘n1’, ‘t1’, ‘sbr_i’, ‘metastasis/relapse’, ‘yes’, ‘positive’] are mapped to ‘1’, indicating positive test results or the presence of certain conditions. The values [‘t2’, ‘n2’, ‘sbr_ii’] are mapped to ‘2’, representing a middle level of a certain condition or stage. The values [‘n3’, ‘t3’, ‘2_to_4’, ‘sbr_iii’] are mapped to ‘3’, likely indicating a more advanced stage of a condition. The value ‘5_or_more’ is mapped to ‘5.5’, providing a continuous scale for certain variables where this applies. The term ‘unknown’ is mapped to NaN, indicating missing or undefined values that may need to be handled during further data processing or model training.
- Handling missing values: handling missing values is crucial for maintaining the integrity of data analysis, and Table 2 illustrates the various techniques used to manage missing data effectively.
Table 2
| Variable | Filling technique |
|---|---|
| Menopause | If is_younger == 1 then fill with 0 |
| If is_younger == 0 then fill with 1 | |
| Number_of_full_term_pregnancies | If nulliparity == 1 then fill with 0 |
| Otherwise fill with the mean value | |
| Nulliparity | If number_of_full_term_pregnancies == 0 then 1 |
| If number_of_full_term_pregnancies != 0 then 0 | |
| Oral_contraception_use; obesity; vascular_invasion | Fill using the random imputation method to keep the distribution |
| Tumor_size; lymph_nodes; sbr_grade | Fill using the mean imputation method because they are quantified features |
| Familial_history_of_breast_cancer; radiotherapy; trastuzumab; adjuvant_chemotherapy | Fill using the mode imputation method because it there is a big difference between the two values in term of frequencies |
| Er; pr; her2 | Fill using the predictive imputation method using Random Forest Classifier model (Converge to a strong relationship between features) |
The previous data cleaning steps were important for transforming the dataset into a usable format for analysis. By dropping irrelevant or incomplete data, creating meaningful binary variables, and converting categorical data into numerical values, the dataset becomes more suitable for ML models. These processes enhance the model’s ability to accurately diagnose breast cancer by providing clean, consistent, and interpretable data. As a result, the following figure (Figure 2) displays the distribution of both numerical and categorical variables. The graphical representation is important, as it provides a clear and intuitive understanding of the data’s underlying patterns, variability, and potential correlations, aiding in more informed decision-making and analysis (30).
Data normalization
Data normalization is important for ensuring that all features contribute equally to the analysis, avoiding biases due to differing scales and improving the performance of ML models (31). To facilitate this, histological_type and surgery_type are converted into new features using the One Hot Encoder technique, and ‘number_of_full_term_pregnancies’, ‘tumor_size’, ‘lymph_nodes’, and ‘sbr_grade’ are normalized using L2 normalization.
Handling imbalanced data
Handling imbalanced data is crucial because it ensures that the model can learn to recognize and predict minority class instances effectively. Without addressing imbalances, models may become biased towards the majority class, leading to inaccurate and skewed predictions, particularly for the underrepresented classes (32).
After completing the pre-processing stage, our dataset became highly imbalanced, potentially leading to a biased model. For instance, if one class significantly outnumbers the others, the model may become overly tuned to the majority class, leading to poor performance on the minority class. This bias results in misleadingly high overall accuracy while failing to capture the nuances of less represented categories. Figure 3 clearly illustrates the imbalance in the dataset, with 23.3% of the data representing one class and 76.7% representing the other. This significant disparity highlights the need for methods to address class imbalance, as the uneven distribution may lead to biased model performance and reduced predictive accuracy for the minority class.
In order to resolve the problem, we have adopted oversampling using the SMOTE, which involves generating synthetic samples for the minority class to balance the dataset, and is specifically utilized when the data is unbalanced to enhance the model’s ability to learn from and predict both classes effectively (33). This step allowed the model to learn more effectively from both classes by addressing class imbalance and creating a more balanced dataset (34). By applying SMOTE in our study, we ensure that synthetic samples are generated based on the most relevant features, improving the overall quality and performance of our models.
Model training and evaluation process
This section outlines the essential processes involved in developing and evaluating ML models. It begins with feature selection, identifying the most relevant features to enhance model performance. A correlation matrix follows, analyzing relationships between variables to uncover interactions and redundancies. We then discuss model training and evaluation, detailing the methodologies for training models and assessing their performance. This leads to the implementation of models and the evaluation of evaluation metrics, which quantify the effectiveness of each model. This approach ensures robust and reliable predictions.
Feature selection
Feature selection techniques are essential in ML to identify the most relevant features in a dataset, enhancing model performance and interpretability while reducing complexity. These techniques help remove redundant or irrelevant features, minimizing overfitting and improving computational efficiency (35). The main approaches to feature selection include filter methods, which use statistical metrics to rank features independently of the model; wrapper methods, which evaluate feature subsets based on model performance (36); and embedded methods, which integrate feature selection within the model training process. By carefully selecting features, ML models can achieve higher accuracy, faster training times, and better generalization to new data (37).
Correlation matrix of dataset
A correlation matrix is a table that shows the pairwise correlation coefficients between multiple variables, helping to identify the strength and direction of relationships between features in a dataset (38). The correlation matrix of our dataset is presented below (Figures 4,5), providing a comprehensive view of the relationships between different features. This matrix reveals how features are correlated with one another, helping to identify potential dependencies and multicollinearity that may influence model performance.
Model training
Model training with the use of performance metrics in ML are crucial for evaluating model effectiveness, especially in breast cancer detection. Additionally, support provided context by indicating the number of actual occurrences of each class. Together, these metrics ensure a detailed and actionable view of model performance, addressing class imbalance and enhancing the reliability of cancer detection results (39).
Models’ implementation and evaluation metrics
To provide a comprehensive evaluation of the models, we calculated the AUC for each method. Additionally, we determined the optimal cut-off points using the Youden Index method (26). From these cut-offs, sensitivity, specificity, positive predictive value (PPV), and negative predictive value (NPV) were computed to give a holistic view of each model’s performance. Class 0 refers to non-cancerous cases (negative class), and class 1 refers to cancerous cases (positive class). Therefore, diagnosing cancer focuses on distinguishing between these two classes. These metrics allow us to assess how well the models distinguish between these two classes, providing insights into both the accuracy of predicting negative cases and the model’s ability to correctly identify cancerous ones.
SVM
The AUC values for the SVM models with different kernel options are presented in Table 3. The linear SVM achieved an AUC of 0.82, the RBF kernel achieved 0.87, and the polynomial kernel achieved 0.79. The optimal cut-off values were determined for each kernel type, and corresponding sensitivity and specificity metrics are provided in Table 3.
Table 3
| Kernel type | AUC | Optimal cut-off | Sensitivity (%) | Specificity (%) | PPV (%) | NPV (%) |
|---|---|---|---|---|---|---|
| Linear | 0.82 | 0.50 | 74.58 | 72.15 | 66.67 | 79.17 |
| RBF | 0.87 | 0.52 | 80.02 | 70.89 | 67.31 | 82.12 |
| Polynomial | 0.79 | 0.48 | 75.11 | 67.02 | 62.45 | 78.54 |
AUC, area under the curve; NPV, negative predictive value; PPV, positive predictive value; RBF, radial basis function; SVM, Support Vector Machine.
The RBF kernel consistently outperformed the other kernels, as indicated by its higher AUC value and improved sensitivity and specificity. These findings align with the need to optimize kernel selection for SVM models.
LR
For the logistic regression model (Table 4), the AUC was calculated as 0.85. The Youden Index method identified an optimal cut-off value of 0.54, resulting in a sensitivity of 79.00% and specificity of 69.00%.
Table 4
| Class | AUC | Optimal cut-off | Sensitivity (%) | Specificity (%) | PPV (%) | NPV (%) |
|---|---|---|---|---|---|---|
| 0 | 0.85 | 0.54 | 79.00 | 69.00 | 67.00 | 81.00 |
AUC, area under the curve; NPV, negative predictive value; PPV, positive predictive value.
These results reflect the balanced performance of the LR model across both classes, highlighting the value of stepwise variable selection in optimizing the model.
RF
The AUC for the RF model was 0.92, showcasing its ability to discriminate between positive and negative classes effectively (Table 5). The Youden Index method identified an optimal cut-off of 0.57, resulting in high specificity (87%) but moderate sensitivity (92%) for the positive class.
Table 5
| Class | AUC | Optimal cut-off | Sensitivity (%) | Specificity (%) | PPV (%) | NPV (%) |
|---|---|---|---|---|---|---|
| 0 | 0.92 | 0.57 | 92 | 87 | 75 | 88 |
AUC, area under the curve; NPV, negative predictive value; PPV, positive predictive value; RF, Random Forest.
The RF model demonstrates strong performance in identifying negative cases but requires further tuning to improve sensitivity for positive cases.
ANN
As illustrated in Table 6, the ANN model achieved an AUC of 0.88. The optimal cut-off, determined at 0.55, resulted in a sensitivity of 90.91% for the negative class and 50.00% for the positive class, with corresponding specificities of 83.33% and 66.67%, respectively.
Table 6
| Class | AUC | Optimal cut-off | Sensitivity (%) | Specificity (%) | PPV (%) | NPV (%) |
|---|---|---|---|---|---|---|
| 0 | 0.88 | 0.55 | 90.91 | 83.33 | 66.67 | 83.33 |
| 1 | 0.88 | 0.55 | 50.00 | 66.67 | 66.67 | 90.91 |
ANN, Artificial Neural Networks; AUC, area under the curve; NPV, negative predictive value; PPV, positive predictive value.
These results indicate the ANN’s robustness in identifying non-cancerous cases but highlight challenges in identifying cancerous ones.
Comparative analysis and discussion
The results of this study underscore the potential of RF and ANN in breast cancer diagnosis, with RF achieving a precision of 84.72%, recall of 92.42%, and an F-score of 88.41%, and ANN achieving a precision of 83.33%, recall of 90.91%, and an F-score of 86.96%. These results demonstrate strong overall performance, particularly in identifying negative cases (class 0). However, both models faced notable challenges in predicting positive cases (class 1), with RF and ANN recall values dropping to 54.17% and 50%, respectively. This limitation, compounded by the relatively small dataset size, has significant implications for the reliability and clinical applicability of the models.
- The small dataset size presents a critical limitation for this study. ML models, particularly complex ones like ANN, require large amounts of data to effectively learn intricate patterns and generalize to new, unseen samples. With a limited number of instances, the models are more prone to overfitting, where they perform well on the training data but struggle to generalize to external datasets. This issue is especially pronounced in medical datasets where variability in patient characteristics and conditions can be substantial. The size of the dataset also exacerbates the challenge of class imbalance. With fewer samples, the minority class (positive cases) is underrepresented, further limiting the model’s ability to accurately learn and predict this critical class. This issue directly impacts recall for positive cases, a crucial metric in clinical applications, as failing to identify true cases of breast cancer (false negatives) can delay diagnosis and treatment.
- The limited dataset size also reduces the statistical reliability of the performance metrics. Precision, recall, and F-scores derived from small datasets may fluctuate significantly with minor changes in the data or preprocessing techniques. This variability underscores the need for caution when interpreting the results and highlights the importance of testing models on larger, more diverse datasets to ensure robustness.
- To mitigate the effects of a small dataset, future research should prioritize the acquisition and use of larger and more diverse datasets. Techniques such as data augmentation, which artificially increases dataset size by generating synthetic samples, can also be explored. This approach is particularly relevant in the medical domain, where collecting extensive datasets may be logistically or ethically challenging. Additionally, cross-validation techniques, such as k-fold cross-validation, should be rigorously applied to maximize the utility of small datasets while minimizing the risk of overfitting. Exploring transfer learning, where models pretrained on large datasets are fine-tuned on smaller, domain-specific datasets, may also provide a pathway to improve performance despite limited data availability.
The findings of this study highlight the potential of RF and ANN in breast cancer diagnosis but underscore the challenges posed by a small dataset, particularly in addressing class imbalance and ensuring generalizability. Future research should focus on leveraging larger datasets, employing data augmentation techniques, and exploring advanced methods like transfer learning to improve model reliability and clinical relevance. Addressing these challenges is critical to advancing ML applications in breast cancer diagnosis and ensuring their effectiveness in real-world settings.
Conclusions
This study highlights the potential of ML techniques, particularly RF and ANN, in breast cancer diagnosis. RF showed robust performance in identifying non-cancerous cases, achieving an AUC of 0.91, while ANN demonstrated high sensitivity for negative cases with an AUC of 0.88. However, both models encountered challenges in accurately predicting cancerous cases, reflecting limitations in recall for the positive class. These challenges are amplified by the small dataset size and class imbalance, which undermine the models’ generalizability and clinical applicability. The limited dataset impacts the robustness of performance metrics like precision and recall, underscoring the need for larger, more diverse datasets in future studies. Addressing class imbalance through advanced preprocessing and leveraging data augmentation techniques can significantly enhance ML models’ effectiveness. This research reinforces the transformative potential of ML in improving breast cancer outcomes while emphasizing the critical need for high-quality data to ensure clinical reliability. Future research should prioritize exploring advanced algorithms and leveraging diverse and larger datasets to enhance model generalizability. Additionally, integrating ensemble techniques and genetic data could further refine predictive accuracy while providing valuable insights into the biological mechanisms of breast cancer.
Acknowledgments
None.
Footnote
Peer Review File: Available at https://jmai.amegroups.com/article/view/10.21037/jmai-24-368/prf
Funding: None.
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://jmai.amegroups.com/article/view/10.21037/jmai-24-368/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Sun YS, Zhao Z, Yang ZN, et al. Risk Factors and Preventions of Breast Cancer. Int J Biol Sci 2017;13:1387-97. [Crossref] [PubMed]
- Guo Y, Zhang H, Yuan L, et al. Machine learning and new insights for breast cancer diagnosis. J Int Med Res 2024;52:3000605241237867. [Crossref] [PubMed]
- Kakushadze Z, Raghubanshi R, Yu W. Estimating cost savings from early cancer diagnosis. Data 2017;2:30. [Crossref]
- Yue W, Wang Z, Chen H, et al. Machine learning with applications in breast cancer diagnosis and prognosis. Designs 2018;2:13. [Crossref]
- Rabiei R, Ayyoubzadeh SM, Sohrabei S, et al. Prediction of Breast Cancer using Machine Learning Approaches. J Biomed Phys Eng 2022;12:297-308. [Crossref] [PubMed]
- Vaka AR, Soni B, Kumar SR. Breast cancer detection by leveraging machine learning. ICT Express 2020;6:320-4. [Crossref]
- Radak M, Lafta HY, Fallahi H. Machine learning and deep learning techniques for breast cancer diagnosis and classification: a comprehensive review of medical imaging studies. J Cancer Res Clin Oncol 2023;149:10473-91. [Crossref] [PubMed]
- Ghorbian M, Ghorbian S. Usefulness of machine learning and deep learning approaches in screening and early detection of breast cancer. Heliyon 2023;9:e22427. [Crossref] [PubMed]
- Hearst MA, Dumais ST, Osuna E, et al. Support vector machines. IEEE Intelligent Systems and their Applications 1998;13:18-28. [Crossref]
- Chen M, Jia Y. Support vector machine-based diagnosis of breast cancer. 2020 International Conference on Communications, Information System and Computer Engineering (CISCE); Kuala Lumpur, Malaysia. IEEE 2020:321-5.
- Akay MF. Support vector machines combined with feature selection for breast cancer diagnosis. Expert Systems with Applications 2009;36:3240-7. [Crossref]
- Al-Salihy NKh, Ibrikci T. Classifying breast cancer using decision tree algorithms. In: Proceedings of the 6th International Conference on Software and Computer Applications. New York, NY, USA: ACM; 2017:144-8. (ICSCA ’17).
- Seo DW, Yi H, Bae HJ, et al. Prediction of Neurologically Intact Survival in Cardiac Arrest Patients without Pre-Hospital Return of Spontaneous Circulation: Machine Learning Approach. J Clin Med 2021;10:1089. [Crossref] [PubMed]
- Minnoor M, Baths V. Diagnosis of breast cancer using random forests. Procedia Comput Sci 2023;218:429-37. [Crossref]
- Susheel Kumar SM, Laxkar D, Adhikari S, et al. Assessment of various supervised learning algorithms using different performance metrics. IOP Conf Ser: Mater Sci Eng 2017;263:042087.
- Rajaguru H. S R SC. Analysis of Decision Tree and K-Nearest Neighbor Algorithm in the Classification of Breast Cancer. Asian Pac J Cancer Prev 2019;20:3777-81. [Crossref] [PubMed]
- Mangukiya M, Vaghani A, Savani M. Breast cancer detection with machine learning. IJRASET 2022;10:141-5. [Crossref]
- Rabiei R, Ayyoubzadeh SM, Sohrabei S, et al. Prediction of Breast Cancer using Machine Learning Approaches. J Biomed Phys Eng 2022;12:297-308. [Crossref] [PubMed]
- Tang Y, Yang B, Peng H, et al. Multi-stages attention breast cancer classification based on nonlinear spiking neural P neurons with autapses. Eng Appl Artif Intell. 2025;142:109869. [Crossref]
- Gupta SR. Prediction time of breast cancer tumor recurrence using Machine Learning. Cancer Treat Res Commun 2022;32:100602. [Crossref] [PubMed]
- Heiberger RM, Holland B. Logistic regression. In: Heiberger RM, Holland B, editors. Statistical Analysis and Data Display: An Intermediate Course with Examples in R. New York, NY: Springer; 2015:593-629.
- Khandezamin Z, Naderan M, Rashti MJ. Detection and classification of breast cancer using logistic regression feature selection and GMDH classifier. J Biomed Inform 2020;111:103591. [Crossref] [PubMed]
- Guo Z, Zhang L, Zhang D. A completed modeling of local binary pattern operator for texture classification. IEEE Trans Image Process 2010;19:1657-63. [Crossref] [PubMed]
- Chawla NV, Bowyer KW, Hall LO, et al. SMOTE: Synthetic minority over-sampling technique. J Artif Intell Res 2002;16:321-57. [Crossref]
- YOUDEN WJ. Index for rating diagnostic tests. Cancer 1950;3:32-5. [Crossref] [PubMed]
- Slaoui M, Mouh FZ, Ghanname I, et al. Outcome of Breast Cancer in Moroccan Young Women Correlated to Clinic-Pathological Features, Risk Factors and Treatment: A Comparative Study of 716 Cases in a Single Institution. PLoS One 2016;11:e0164841. [Crossref] [PubMed]
- Mohammed SA, Darrab S, Noaman SA, et al. Analysis of breast cancer detection using different machine learning techniques. In: Tan Y, Shi Y, Tuba M, editors. Data Mining and Big Data. Singapore: Springer; 2020:108-17.
- Berisha V, Krantsevich C, Hahn PR, et al. Digital medicine and the curse of dimensionality. NPJ Digit Med 2021;4:153. [Crossref] [PubMed]
- Nguyen THT, Dinh DT, Sriboonchitta S, et al. A method for k-means-like clustering of categorical data. J Ambient Intell Hum Comput 2023;14:15011-21. [Crossref]
- Etchegaray JM, Fischer WG. Understanding evidence-based research methods: graphical data analysis. HERD 2010;3:118-25. [Crossref] [PubMed]
- de Amorim LBV, Cavalcanti GDC, Cruz RMO. The choice of scaling technique matters for classification performance. Appl Soft Comput 2023;133:109924. [Crossref]
- Mulugeta G, Zewotir T, Tegegne AS, et al. Classification of imbalanced data using machine learning algorithms to predict the risk of renal graft failures in Ethiopia. BMC Med Inform Decis Mak 2023;23:98. [Crossref] [PubMed]
- Assegie TA, Salau AO, Sampath K, et al. Evaluation of adaptive synthetic resampling technique for imbalanced breast cancer identification. Procedia Comput Sci 2024;235:1000-7. [Crossref]
- Wongvorachan T, He S, Bulut O. A comparison of undersampling, oversampling, and SMOTE methods for dealing with imbalanced classification in educational data mining. Information 2023;14:54. [Crossref]
- Mamdouh Farghaly H, Abd El-Hafeez T. A high-quality feature selection method based on frequent and correlated items for text classification. Soft Comput 2023;27:11259-74. [Crossref]
- Parr T, Hamrick J, Wilson JD. Nonparametric feature impact and importance. Inf Sci 2024;653:119563. [Crossref]
- Koirunnisa K, Siregar AM, Faisal S. Optimized machine learning performance with feature selection for breast cancer disease classification. Jurnal Ilmiah Teknik Elektro Komputer dan Informatika 2023;9:1131-43.
- Temesi J, Szádoczki Z, Bozóki S. Incomplete pairwise comparison matrices: Ranking top women tennis players. J Oper Res Soc 2024;75:145-57. [Crossref]
- McKinney SM, Sieniek M, Godbole V, et al. International evaluation of an AI system for breast cancer screening. Nature 2020;577:89-94. [Crossref] [PubMed]
Cite this article as: Tachicart R, Rezki H, Korchi A, Abatal A. A comparative study of machine learning algorithms for breast cancer diagnosis. J Med Artif Intell 2025;8:58.






