
Pediatric pneumonia X-ray image classification: predictive model development with DenseNet-169 transfer learning
Highlight box
Key findings
• The study demonstrated that deep learning, particularly convolutional neural networks (CNNs), has the potential to significantly enhance the early diagnosis and treatment of pneumonia. This can lead to improved pediatric patient outcomes by providing more accurate and efficient diagnostic methods.
• Transfer learning using the DenseNet-169 CNN on the pneumonia dataset achieved a high accuracy of 91.6% in classifying X-ray images as normal or pneumonia, offering fast and reliable diagnostic approach while reducing the likelihood of human error.
What is known and what is new?
• Conventional pneumonia diagnostic methods, including chest X-rays, heavily rely on the expertise of medical professionals, often resulting in inconsistencies and misdiagnoses, particularly in resource-limited settings.
• This study presents an artificial intelligence (AI)-driven methodology utilizing CNNs to evaluate chest X-ray images, streamlining the detection process with enhanced accuracy and speed for pneumonia identification.
What is the implication, and what should change now?
• Integrating AI-powered tools into clinical practice for diagnosing pneumonia has the potential to enable earlier detection and treatment, therefore reducing mortality rates and improving the quality of patient care globally.
• There is a need to refine AI models further, to ensure their reliability and applicability across diverse populations. Additionally, addressing ethical and regulatory concerns is crucial for the effective implementation of AI in healthcare.
Introduction
Pneumonia, a serious infection of the lungs, results from various microbial pathogens, leading to inflammation and the accumulation of pus and fluid in the alveoli. This compromises the exchange of oxygen and carbon dioxide, posing significant global health challenges, especially for vulnerable populations like infants, the elderly, and immunocompromised individuals (1). With approximately 4 million deaths annually, pneumonia remains one of the leading causes of mortality worldwide (2,3). The condition is caused by bacteria, viruses, or fungi, necessitating distinct treatment strategies. Early and accurate diagnosis is critical to reducing pneumonia-related morbidity and mortality (4,5), as timely detection enables the isolation of affected patients and the initiation of targeted therapy, mitigating the rapid spread of infection and its complications (6).
Traditional diagnostic methods, including clinical examination, blood tests, and chest X-rays, rely heavily on clinician expertise but are limited by subjective variability and the potential for misdiagnosis. For example, the diagnostic accuracy of chest X-rays ranges between 70% and 80%, depending on the clinician’s experience and the complexity of cases (7,8). Moreover, resource-limited settings often lack skilled radiologists, further hindering timely and accurate diagnosis. In contrast, deep learning (DL) approaches have shown promise in automating pneumonia detection from chest X-ray images, achieving diagnostic accuracies exceeding 90% on benchmark datasets (9-14). These technologies can enhance diagnostic efficiency by addressing the overlapping features in chest radiographs of pneumonia and normal, which can otherwise lead to clinician misinterpretation (15).
DL has revolutionized medical imaging by automating complex diagnostic tasks like image segmentation, lesion detection, and disease classification (16-19). Its adoption in clinical diagnostics is motivated by improved precision, reduced manual workload, and faster analysis (20-22). However, significant challenges remain, including model generalizability across diverse populations, imaging protocols, and data quality. The interpretability of DL models also raises concerns, as clinicians require transparency in decision-making (23,24). Ethical issues, including patient privacy and compliance with healthcare regulations, further complicate the integration of these technologies into clinical workflows (25).
Although extensive research has been conducted on pneumonia detection using DL, studies focusing on Pediatric populations remain limited. Additionally, imbalanced datasets often hinder model performance, highlighting the need for advanced data augmentation techniques to ensure robustness. By addressing these challenges, this study aims to contribute valuable insights to the field of AI-assisted medical imaging. We present this article in accordance with the TRIPOD reporting checklist (available at https://jmai.amegroups.com/article/view/10.21037/jmai-24-356/rc).
Methods
Dataset
The dataset used in this study, sourced from Kaggle, consists of 5,866 chest X-ray images categorized into two groups: pneumonia and normal (sample images shown in Figure 1). These images were organized into three primary folders: train, validation (val), and test. The input dataset comprises a training set with 5,266 images accounting for 89.81% of the total, a validation set with 16 images representing 0.27% and a test set with 624 images, making up 9.92%. After preprocessing, the dataset was modified, resulting in a training set of 5,034 images (85.88%), a validation set of 208 images (4.2%) and the test set remaining unchanged at 624 (9.92%). This division ensured a balanced representation of both categories, with no missing or incomplete data, as the Kaggle dataset was rigorously curated and underwent stringent quality control by its providers.
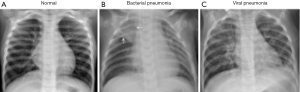
The Kaggle dataset was selected for its comprehensive and high-quality collection of annotated chest X-ray images, making it particularly suitable for training DL models for pneumonia detection. The dataset includes both positive (pneumonia) and negative (normal) cases that were curated from pediatric patients aged 1 to 5 years, a demographic highly susceptible to pneumonia due to their still-developing immune systems. The availability of high-quality anterior-posterior chest X-rays from this age group ensures consistency in analysis and provides a dataset tailored to address the unique physiological characteristics of young children’s lungs. Additionally, the dataset underwent rigorous quality control, with poor-quality or unreadable scans excluded during curation. Diagnoses were graded by two expert physicians, and any discrepancies were resolved through a third expert’s review. This ensured the reliability of labels and enhanced the dataset’s overall quality and relevance.
The dataset (26) used in this study adhered to specific inclusion and exclusion criteria to ensure quality and reliability. For inclusion, the dataset exclusively comprised two distinct image categories—pneumonia and normal—each stored in separate subfolders within the main directories for training, testing, and validation. It contained only anterior-posterior chest X-rays from pediatric patients aged one to five years, sourced from Guangzhou Women and Children’s Medical Center, Guangzhou. To maintain quality, only high-quality and readable scans, as determined by an initial quality control process, were included. The exclusion criteria aimed to enhance reliability and accuracy. Images that were blurry, incomplete, or unreadable were excluded during preprocessing. Furthermore, diagnoses were initially graded by two expert physicians, and in cases of discrepancies, a third expert conducted a review to ensure consistent labeling. Any images with unclear or inconsistent diagnoses were excluded from the dataset. A flowchart regarding exclusion criteria is shown in Figure 2.
Preprocessing and dataset integrity
The Kaggle dataset underwent thorough preprocessing and quality control by its providers, ensuring all included images were suitable for analysis. No additional exclusions were necessary during this study. Since the dataset was complete and ready for use, techniques such as interpolation or imputation to address missing data were not required. A detailed number of images per category for training, validation and test sets are shown in Table 1.
Table 1
| Dataset | Total cases | Pneumonia cases | Normal cases |
|---|---|---|---|
| Before preprocessing | |||
| Training set | 5,266 | 3,885 | 1,341 |
| Validation set | 16 | 8 | 8 |
| Test set | 624 | 390 | 234 |
| After preprocessing | |||
| Training set | 5,034 | 3,789 | 1,245 |
| Validation set | 208 | 104 | 104 |
| Test set | 624 | 390 | 234 |
Data augmentation
The data augmentation techniques applied to all images uniformly included several transformations to enhance the dataset. A shear transformation with a range of 0.2 was implemented to simulate geometric distortions, while a zoom transformation of the same range introduced scale variability. Horizontal and vertical shifts were applied, allowing up to 10% (0.1) of the image dimensions to vary. Brightness adjustments were made, ranging between 0.2 and 1.0, to account for differences in illumination. Additionally, horizontal flipping was performed to generate mirrored versions of the images, further diversifying the dataset. Following these augmentations, the dataset expanded to 75,360 augmented training images, calculated as steps_per_epoch × batch_size × epochs =314×16×15. These augmentations ensured consistency in preserving the original data distribution while enhancing the model’s ability to generalize to new, unseen data. Data augmentation significantly increased the diversity of the training set, helping to prevent overfitting and improve the model’s generalization to real-world scenarios. By simulating variations in imaging conditions, such as lighting and orientation, the augmentation process prepared the model to handle a wide range of diagnostic challenges, thereby enhancing its robustness and reliability.
Model training and evaluation
The convolutional neural network (CNN) stands out as the primary DL algorithm extensively employed for image classification tasks (27). CNNs consist of a convolutional layer, a pooling layer, a non-linear layer, and fully connected layers (28). A deep neural network (DNN) architecture uses several neurons to extract significant features from images. To effectively build and train a neural network from the scratch, a substantial amount of data is necessary to ensure it learns features adequately without succumbing to overfitting/underfitting. Building a neural network that can fit to smaller dataset is challenging as initial weights are randomly initialized and updated continuously based on the dataset and the loss function used. This can result in underfitting the network due to insufficient training data and also time-consuming due to slow updates to the weights during the learning process. There are various state-of-the-art CNN models used for the classification of images such as VGG (29), ResNet (30), GoogleNet (31), DenseNet (32), etc. used for classification in various sectors.
This paper presents a study of Pneumonia classification using the transfer learning of DenseNet-169 shown in Figure 3 (33). Transfer learning is a technique used in the training of DL models that makes use of pre-trained CNN weights trained on state-of-the-art datasets. During the transfer learning the higher-level representations from the new dataset are only trained while transferring the lower-level representations. Thus, transfer learning involves adjusting the weights of higher hidden layers and the effectiveness of learning is determined using the distance between origin and destination datasets. Despite the large number of layers compared to other State-Of-The-Art classification models, DenseNet has a relatively low number of parameters handling the vanishing gradient problem. In this work, a DenseNet-169 classification network pre-trained on the ImageNet dataset is fine-tuned using the pneumonia dataset through transfer learning. To adapt DenseNet-169 for a binary classification task, the original top (or head) of the model was replaced with a custom layer structure tailored to the binary classification of Pneumonia as shown in Table 2. Like many pre-trained models, DenseNet-169 is designed for multi-class classification task, for examples ImageNet classification with 1,000 classes. However, for binary classification, the head model requires significant modifications such as removal of the original head. Instead of using traditional fully connected layers, a global average pooling (GAP) layer was added for reducing the spatial dimensions of feature maps mitigating overfitting. A dense layer with a reduced number of neuros was added as bottleneck for condensing the extracted features into a lower-dimensional space with ReLU activation layer to introduce non-linearity and enhance feature learning. A dropout layer was included to prevent overfitting by randomly setting a fraction of inputs to zero during training, making sure that the model does not rely heavily on any single feature. Lastly, the final dense layer was designed with a single neuron with sigmoid activation which outputs probabilities between 0 and 1. This custom head is integrated with the pre-trained DenseNet-169 backbone.
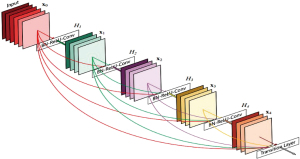
Table 2
| Layer (type) | Output shape | Param # |
|---|---|---|
| densenet169 (functional) | (None, 7, 7, 1,664) | 1, 26, 42,880 |
| global_average_pooling2d | (None, 1,664) | 0 |
| batch_normalization | (None, 1,664) | 6,656 |
| dense | (None, 256) | 4, 26,240 |
| dropout | (None, 256) | 0 |
| batch_normalization_1 | (None, 256) | 1,024 |
| dense_1 | (None, 128) | 32,896 |
| dropout_1 | (None, 128) | 0 |
| dense_2 | (None, 1) | 129 |
Total params: 13,109,825 (50.01 MB). Trainable params: 463,105 (1.77 MB). Non-trainable params: 12,646,720 (48.24 MB).
To align with input requirements of DenseNet-169, images are resized to 224×224. Further, to match the model’s expectation, the images are normalized and standardized during the preprocessing. To mitigate overfitting and enhance the diversity in the dataset, augmentation techniques described in the dataset section were performed. During fine-tuning, the model is initialized using pre-trained weights derived from ImageNet dataset. The initial layers were left frozen to retain generic features that are transferable across domains. The top fully connected layer was replaced to perform pneumonia classification, which includes a classification head comprising of a GAP layer, a dense layer with 128 neurons and ReLU activation followed by a dropout layer and finally a softmax layer with two output units corresponding to normal and pneumonia classes. Thus, transfer learning allows leveraging the existing knowledge of the neural network for the classification of the new dataset on which the transfer learning has been performed. Different hyperparameter combinations are experimented with to find the optimal parameters. The training process involves running the model through 25 epochs, employing a learning rate of 0.0001 along with cross-entropy loss and stochastic gradient descent (SGD) optimizer. A lower learning rate was used for fine-tuning to avoid manipulating pre-trained weights while adapting to the new dataset. This ensures the continual minimization of the loss function and updates the weights after each epoch. A batch size of 16 is chosen to balance computational efficiency and convergence of model with adam optimizer to dynamically adjust the learning rate. The model performance is evaluated on the validation dataset after every epoch to monitor the overfitting. The fine-tuned DenseNet-169 model is evaluated on unseen test data. Test images traverse the network, yielding predictions compared against ground truth labels. Performance metrics including accuracy, precision, recall, and F1-score are calculated to assess model efficiency.
Statistical analysis
Accuracy: this metric quantifies the overall correctness of predictions, representing the ratio of correctly classified samples to the total number of samples.
Precision: precision measures the reliability of positive predictions, indicating the proportion of true positives (TPs) among all instances predicted as positive.
Recall: also known as sensitivity, recall evaluates the model’s ability to capture all relevant instances of the positive class.
F1-score: the F1-score combines precision and recall into a single metric, providing a balanced measure that considers both false positives (FPs) and false negatives (FNs).
The statistical analysis was conducted using Python and other python libraries such as SciPy, Statsmodels. These tools were employed for calculating performance metrics conducting comparative tests and visualizing results. Confidence intervals (CIs) for accuracy, precision, recall and F1-score were computed using standard error-based methods to assess the reliability of the metrics.
The code implementation for the DenseNet-169 model, including preprocessing steps, training, evaluation is available through a publicly accessible google colab link (https://colab.research.google.com/drive/1mRjCy4f5eQhTClH5e3AqMCptRut_4KgI?usp=sharing).
Results
In this section, we present a comprehensive analysis of the performance of our neural network model, focusing on learning curves, confusion matrix evaluation, and predicted results. These components collectively offer insights into the model’s training dynamics, classification accuracy, and specific instances of correct and incorrect predictions. The results of the proposed model are estimated using evaluation metrics such as accuracy, precision, recall, and F1-score on test data as shown in Table 3.
Table 3
| Model | Accuracy (%) (95% CI) | Precision (%) (95% CI) | Recall (%) (95% CI) | F1-score (%) (95% CI) |
|---|---|---|---|---|
| DenseNet-169 | 91.66 (89.37–93.95) | 90.99 (88.62–93.36) | 86.32 (83.50–89.14) | 87.62 (84.96–90.28) |
| DenseNet-121 | 90.41 | 89.25 | 86.73 | 87.97 |
| VGG16 | 76.63 | 82.60 | 76.56 | 79.46 |
CI, confidence interval.
The evaluation of the proposed model was conducted using several standard metrics, including accuracy, precision, recall, and F1-score, based on a large and representative sample of the dataset. These metrics were computed from the classification of both positive and negative instances in relation to the true and false classes. Specifically, TP refers to samples classified as positive that belong to the positive class, while FP refers to samples from the negative class misclassified as positive. True negative (TN) refers to samples from the negative class correctly classified as negative, and FN refers to positive samples that were misclassified as negative. Learning curves shown in Figure 4A depict the model’s performance metrics over epochs, illustrating its learning process on both the training and validation datasets. The training curve demonstrates the model’s ability to fit the training data, ideally showing improvement or stabilization.
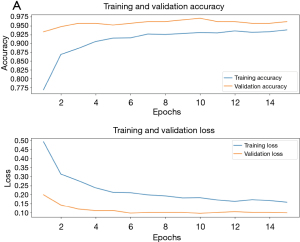
Concurrently, the validation curve indicates the model’s generalization performance, crucial for identifying overfitting or underfitting tendencies. Analysis of these curves informs adjustments to training strategies such as data augmentation, regularization techniques, or model architecture modifications. The learning curves show the fine-tuning of the DenseNet-169 model converges at around 11 epochs. The closeness of the curves for the train and validation set indicates the generalization of the model. This outcome is indicative of the model’s resilience to overfitting, aided by the use of regularization techniques like early stopping. The alignment between training and validation performance underscores the suitability of the selected architecture and hyperparameter configurations for this dataset. The model is then evaluated by passing an unseen test dataset for prediction. The model has an accuracy of 91.66% prediction on unseen data which is shown in Figure 4B by plotting the confusion matrix. The confusion matrix offers a comprehensive overview of how well the model performs in classifying different categories. The actual and predicted labels for sample images from the test dataset are shown in Figure 5. Examining the model’s predictions against ground truth labels offers a granular view of its performance in individual instances.
Discussion
The DenseNet-169 model demonstrated exceptional performance in the classification of pneumonia X-ray images, achieving the highest accuracy of 91.66% compared to DenseNet-121 (90.41%) and VGG-16 (76.63%). Evaluation metrics such as precision (90.99%), recall (86.32%), and F1-score (87.62%) collectively affirm its robustness and effectiveness in identifying Pneumonia cases. The model’s dense connectivity pattern contributed significantly to mitigating vanishing gradient issues and improving parameter efficiency, enabling better feature extraction and gradient flow.
DenseNet-169’s superior architecture, with its deeper layers and dense connectivity, facilitated improved feature reuse and reduced computational redundancy. These characteristics were instrumental in achieving high performance even in the presence of moderate dataset imbalances. Its precision score underscores its ability to minimize false positives, a critical requirement in medical diagnostics where incorrect predictions can have severe implications. The study noted a relatively lower recall compared to precision, highlighting a trade-off in capturing all relevant instances of the positive class. This limitation indicates that some true Pneumonia cases might not be detected, warranting further optimization of the model. Additionally, the dataset used in this study was moderately imbalanced, with a higher proportion of Pneumonia cases. While prior research (34) suggests that such imbalance may not significantly affect accuracy, addressing this imbalance more comprehensively could further enhance recall and overall model reliability.
Compared to other CNN architectures such as DenseNet-121 (35) and VGG-16, DenseNet-169 (36) exhibited superior performance. The higher accuracy and precision of DenseNet-169 can be attributed to its advanced architecture, which enhances gradient flow and facilitates feature reuse (37). In contrast, the relatively lower performance of VGG-16 reflects its less sophisticated design and lack of dense connectivity. These findings align with trends in similar research, where deeper and more connected architectures outperform traditional models in complex image classification tasks. The enhanced performance of DenseNet-169 is primarily due to its dense connectivity pattern, which promotes efficient feature reuse and mitigates vanishing gradient problems. This architectural advantage allowed the model to effectively leverage the limited dataset, extracting meaningful features even from underrepresented patterns. The misclassifications observed in the confusion matrix provide insights into potential areas for improvement, such as augmenting underrepresented classes or refining hyperparameters.
The findings of this study underscore the importance of leveraging advanced DL architectures in medical image classification. Future work should focus on improving recall through targeted data augmentation strategies, enhancing dataset diversity, or incorporating ensemble learning techniques. Addressing dataset imbalances and investigating misclassified instances in greater detail could also contribute to refining the model’s performance. These efforts will be crucial in advancing the reliability and adaptability of AI models in critical healthcare applications.
Conclusions
In summary, this research highlights the effectiveness of DenseNet-169 in pneumonia classification, specific to pediatric patients with pneumonia, attaining an impressive accuracy rate of 91.66%. Utilizing its advanced DL framework, DenseNet-169 has shown significant capability in differentiating pneumonia cases from normal ones based on medical X-ray images. This strong performance underscores its potential as a crucial resource in clinical environments, providing dependable assistance to healthcare practitioners for prompt and precise diagnoses. Future investigations may focus on improving the model’s generalization and scalability, to incorporate these enhancements into practical healthcare applications to better patient outcomes.
Acknowledgments
None.
Footnote
Reporting Checklist: The authors have completed the TRIPOD reporting checklist. Available at https://jmai.amegroups.com/article/view/10.21037/jmai-24-356/rc
Peer Review File: Available at https://jmai.amegroups.com/article/view/10.21037/jmai-24-356/prf
Funding: None.
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://jmai.amegroups.com/article/view/10.21037/jmai-24-356/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted using publicly available, open-source data that was originally collected following ethical guidelines. No new human data was collected, and therefore, additional ethics approval was not required for this study.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Zhang ZX, Yong Y, Tan WC, et al. Prognostic factors for mortality due to pneumonia among adults from different age groups in Singapore and mortality predictions based on PSI and CURB-65. Singapore Med J 2018;59:190-8. [Crossref] [PubMed]
- Bjarnason A, Westin J, Lindh M, et al. Incidence, Etiology, and Outcomes of Community-Acquired Pneumonia: A Population-Based Study. Open Forum Infect Dis 2018;5:ofy010. [Crossref] [PubMed]
- Eurich DT, Marrie TJ, Minhas-Sandhu JK, et al. Risk of heart failure after community acquired pneumonia: prospective controlled study with 10 years of follow-up. BMJ 2017;356:j413. [Crossref] [PubMed]
- Metlay JP, Fine MJ. Testing strategies in the initial management of patients with community-acquired pneumonia. Ann Intern Med 2003;138:109-18. [Crossref] [PubMed]
- Mandell LA. Community-acquired pneumonia: An overview. Postgrad Med 2015;127:607-15. [Crossref] [PubMed]
- Eurich DT, Marrie TJ, Minhas-Sandhu JK, et al. Ten-Year Mortality after Community-acquired Pneumonia. A Prospective Cohort. Am J Respir Crit Care Med 2015;192:597-604. [Crossref] [PubMed]
- Mabrouk A, Díaz Redondo RP, Dahou A, et al. Pneumonia Detection on Chest X-ray Images Using Ensemble of Deep Convolutional Neural Networks. Appl Sci 2022;12:6448. [Crossref]
- Bourcier JE, Paquet J, Seinger M, et al. Performance comparison of lung ultrasound and chest x-ray for the diagnosis of pneumonia in the ED. Am J Emerg Med 2014;32:115-8. [Crossref] [PubMed]
- Han Y, Chen C, Tewfik A, et al. Pneumonia detection on chest X-ray using radiomic features and contrastive learning. Proc IEEE Int Symp Biomed Imaging 2021;2021:247-51. [Crossref] [PubMed]
- Jain DK, Singh T, Saurabh P, et al. Deep Learning-Aided Automated Pneumonia Detection and Classification Using CXR Scans. Comput Intell Neurosci 2022;2022:7474304. [Crossref] [PubMed]
- Sharma S, Guleria K. A deep learning based model for the detection of pneumonia from chest X-ray images using VGG-16 and neural networks. Procedia Comput Sci 2023;218:357-66. [Crossref]
- Stephen O, Sain M, Maduh UJ, et al. An Efficient Deep Learning Approach to Pneumonia Classification in Healthcare. J Healthc Eng 2019;2019:4180949. [Crossref] [PubMed]
- Nillmani, Jain PK, Sharma N, et al. Four Types of Multiclass Frameworks for Pneumonia Classification and Its Validation in X-ray Scans Using Seven Types of Deep Learning Artificial Intelligence Models. Diagnostics (Basel) 2022;12:652.
- Hasan MR, Ullah SMA, Isla SMR. Recent advancement of deep learning techniques for pneumonia prediction from chest X-ray image. Med Rec 2024;7:100106. [Crossref]
- Scanlon GT, Unger JD. The radiology of bacterial and viral pneumonias. Radiol Clin North Am 1973;11:317-38. [Crossref] [PubMed]
- Litjens G, Kooi T, Bejnordi BE, et al. A survey on deep learning in medical image analysis. Med Image Anal 2017;42:60-88. [Crossref] [PubMed]
- Gore JC. Artificial intelligence in medical imaging. Magn Reson Imaging 2020;68:A1-4. [Crossref] [PubMed]
- Sistaninejhad B, Rasi H, Nayeri P. A Review Paper about Deep Learning for Medical Image Analysis. Comput Math Methods Med 2023;2023:7091301. [Crossref] [PubMed]
- Alzubaidi L, Zhang J, Humaidi AJ, et al. Review of deep learning: concepts, CNN architectures, challenges, applications, future directions. J Big Data 2021;8:53. [Crossref] [PubMed]
- Mall PK, Singh PK, Srivastava S, et al. A comprehensive review of deep neural networks for medical image processing: Recent developments and future opportunities. Healthcare Analytics 2023;4:100216. [Crossref]
- Cheung HMC, Rubin D. Challenges and opportunities for artificial intelligence in oncological imaging. Clin Radiol 2021;76:728-36. [Crossref] [PubMed]
- Siegersma KR, Leiner T, Chew DP, et al. Artificial intelligence in cardiovascular imaging: state of the art and implications for the imaging cardiologist. Neth Heart J 2019;27:403-13. [Crossref] [PubMed]
- Dhar T, Dey N, Borra S, et al. Challenges of Deep Learning in Medical Image Analysis—Improving Explainability and Trust. IEEE Trans on Tech Soc 2023;4:68-75. [Crossref]
- Borys K, Schmitt YA, Nauta M, et al. Explainable AI in medical imaging: An overview for clinical practitioners - Beyond saliency-based XAI approaches. Eur J Radiol 2023;162:110786. [Crossref] [PubMed]
- Naik N, Hameed BMZ, Shetty DK, et al. Legal and Ethical Consideration in Artificial Intelligence in Healthcare: Who Takes Responsibility? Front Surg 2022;9:862322. [Crossref] [PubMed]
- Kermany D, Zhang K, Goldbaum M. Labeled Optical Coherence Tomography (OCT) and Chest X-Ray Images for Classification. Mendeley Data 2018; [Crossref]
- LeCun Y, Bengio Y, Hinton G. Deep learning. Nature 2015;521:436-44. [Crossref] [PubMed]
- Albawi S, Mohammed TA, Al-Zawi S. Understanding of a convolutional neural network. International Conference on Engineering and Technology (ICET) 2017:1-6. doi:
10.1109/ICEngTechnol.2017.8308186 .10.1109/ICEngTechnol.2017.8308186 - Simonyan K, Zisserman A. Very Deep Convolutional Networks for Large-Scale Image Recognition. 3rd International Conference on Learning Representations (ICLR) 2015:1-14.
- He K, Zhang X, Ren S, Sun J. Deep Residual Learning for Image Recognition. IEEE Conference on Computer Vision and Pattern Recognition (CVPR) 2016:770-8.
- Szegedy C, Liu W, Jia Y, et al. Going Deeper with Convolutions. Proc IEEE Comput Soc Conf Comput Vis Pattern Recognit 2015;1-9.
- Huang G, Liu Z, van der Maaten L, et al. Densely Connected Convolutional Networks. Proc IEEE Comput Soc Conf Comput Vis Pattern Recognit 2017;4700-8.
- Ribani R, Marengoni M. A Survey of Transfer Learning for Convolutional Neural Networks. 32nd SIBGRAPI Conference on Graphics, Patterns and Images Tutorials (SIBGRAPI-T) 2019:47-57.
- Davidian M, Lahav A, Joshua BZ, et al. Exploring the Interplay of Dataset Size and Imbalance on CNN Performance in Healthcare: Using X-rays to Identify COVID-19 Patients. Diagnostics (Basel) 2024;14:1727. [Crossref] [PubMed]
- Rajpurkar P, Irvin J, Ball RL, et al. Deep learning for chest radiograph diagnosis: A retrospective comparison of the CheXNeXt algorithm to practicing radiologists. PLoS Med 2018;15:e1002686. [Crossref] [PubMed]
- Wang X, Peng Y, Lu L, et al. ChestX-ray8: Hospital-scale Chest X-ray Database and Benchmarks on Weakly-supervised Classification and Localization of Common Thorax Diseases. IEEE Conference on Computer Vision and Pattern Recognition (CVPR) 2017:3462-71.
- Kermany DS, Goldbaum M, Cai W, et al. Identifying Medical Diagnoses and Treatable Diseases by Image-Based Deep Learning. Cell 2018;172:1122-1131.e9. [Crossref] [PubMed]
Cite this article as: Katreddi S, Midatani A, Roy AP, Velpuri U, Kasani S. Pediatric pneumonia X-ray image classification: predictive model development with DenseNet-169 transfer learning. J Med Artif Intell 2025;8:37.




