
Deploy your own machine learning model with no coding expertise: resident perspective
Introduction
Most machine learning (ML) research projects involve a collaborative approach, where teams often consist of data scientists and domain experts. Data scientists typically possess expertise in coding languages like Python, while doctors contribute domain-specific knowledge, such as recognizing medical lesions and understanding clinical needs for specific model outcomes. As residents without coding expertise, we had numerous ideas about how artificial intelligence (AI) could be leveraged to address various clinical needs. However, most ML models are traditionally trained using coding languages like Python or R.
With the advent of automatic ML (AutoML) platforms, it has become easier to create and implement AI models without coding knowledge. One such platform is Google Cloud’s AutoML (Vertex AI), which allows users to either use a pre-trained model or create a custom model (1). Researchers can upload their own datasets, label or leave images unlabeled through an intuitive interface, train the model, and subsequently use it to make predictions.
The aim of this study was to develop a ML model (polyp detector) using a no-code approach, making it accessible for clinical application without requiring coding expertise. This methodology can be used by a student or resident to test proof their concepts by building a ML model.
Methodology
Process of training a ML model without coding
Step 1: selecting platform
There are several platforms (e.g., Google AutoML, IBM Watson, Microsoft Azure Cognitive Services) that can be used to create a ML model without coding. For this research, Google’s AutoML Image was utilized, which employs ML to analyze the content of image data. AutoML allows for the training of an ML model to classify image data or detect objects within images.
Step 2: importing data
The image dataset can be uploaded from a computer to the online platform. Once the images are preprocessed, they are stored in a new cloud storage bucket.
Step 3: classification
The dataset is divided into training, validation, and test sets manually or by default. Google Vertex AI automatically allocates 80% of the images for training, 10% for validation, and 10% for testing.
Step 4: training
Authors individually labeled the polyps in each image. Once the images are labeled, the model was trained. During this process, the model is both validated and tested automatically.
Step 5: evaluation
After training, the model’s performance is evaluated based on the data provided. If needed, additional data can be introduced to improve metrics such as sensitivity (recall) or positive predictive value (precision).
Step 6: deployment
To enable the model to predict new data, endpoints must be created. Once the model is deployed to an endpoint, it can be used to test new images or generate batch predictions. An email link is provided to the user, which can be shared to allow others access to the model’s predictions.
Step 7: testing
The deployed model can be used to process new datasets. Users can upload single images or batches to assess the model’s performance. Each image processed by the model is accompanied by a confidence score, which represents the probability (ranging from 0 to 1) that the object within the bounding box is correctly classified.
Results/demo
Polyp detector
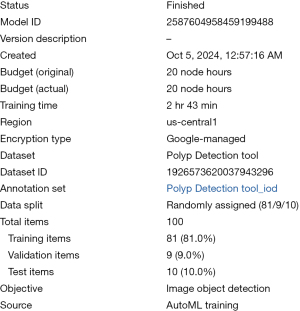
We used 100 colonoscopy images containing 110 polyps from the publicly available Kvasir dataset (2). Images were uploaded into AutoML (Google Cloud’s Vertex AI) on the Google Cloud platform. Once the images were preprocessed and stored on the cloud, the model was trained. Since AutoML is an automated platform, the user only needs to wait for a few hours until the trained model is ready. The dataset used was the polyp detection tool dataset (dataset ID: 1926573620037943296), consisting of 100 images divided randomly into 81% training (81 items), 9% validation (9 items), and 10% testing (10 items).
Once the model was trained, it was deployed to an endpoint for use on new data. The polyp detection achieved an average precision of 100%, a precision of 64.5%, and a recall of 54.5% (Figure 1). After deployment, the model was tested with new images that had never been processed by the platform before, and it successfully detected polyps in all of them (Figure 1).
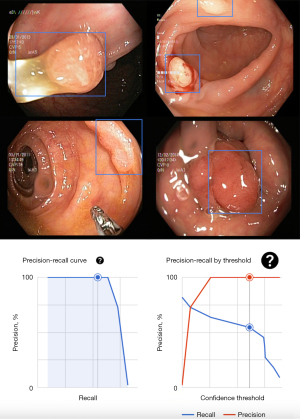
The AutoML model for polyp detection, created using Google Cloud’s Vertex AI, was completed on October 5, 2024, with a total training time of 2 hours and 43 minutes using 20 node hours. The model, identified by model ID 2587604958459199488, was developed in the us-central1 region under a Google-managed encryption type (Figure 2).
Limitations
One of the primary limitations of AutoML is its lack of flexibility and customization compared to traditional ML models. While AutoML simplifies the model development process, it restricts users from fine-tuning hyperparameters, optimizing model architectures, or incorporating domain-specific modifications, which can sometimes lead to suboptimal performance. Another major limitation is the requirement for high-quality labeled data. AutoML platforms heavily rely on the quality and diversity of training data, and biases in the dataset can directly impact the model’s accuracy and generalizability. Furthermore, AutoML is often a black-box approach, meaning that users have limited visibility into the inner workings of the model, making it difficult to interpret predictions and troubleshoot errors. This lack of explainability can be a concern in high-stakes applications like healthcare, where understanding model decisions is crucial.
Conclusions
This paper explores AutoMLs like Google Cloud’s Vertex AI, enabling clinicians with no coding expertise to create and deploy ML models. By utilizing a no-code approach, the model was trained, validated, and deployed for real-time clinical application. The results demonstrated the effectiveness of AutoML in building an efficient ML model. This approach highlights how non-coders can easily leverage these platforms for healthcare solutions.
Acknowledgments
None.
Footnote
Peer Review File: Available at https://jmai.amegroups.com/article/view/10.21037/jmai-24-448/prf
Funding: None.
Conflicts of Interest: Both authors have completed the ICMJE uniform disclosure form (available at https://jmai.amegroups.com/article/view/10.21037/jmai-24-448/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Google. (n.d.). Automl Beginner’s guide | vertex AI | google cloud. Google. Available online: https://cloud.google.com/vertex-ai/docs/beginner/beginners-guide [access date: 10/04/2024]
- Pogorelov K, Randel KR, Griwodz C, et al. KVASIR: A Multi-Class Image Dataset for Computer Aided Gastrointestinal Disease Detection. In: Proceedings of the 8th ACM on Multimedia Systems Conference (MMSys'17). New York, NY, USA: ACM; 2017:164-9. doi:
10.1145/3083187.3083212 .10.1145/3083187.3083212
Cite this article as: Singh J, Agrawal A. Deploy your own machine learning model with no coding expertise: resident perspective. J Med Artif Intell 2025;8:50.


