
Smartphone-based polyp detection: a first step towards an open-source AI framework
Introduction
Colorectal cancer (CRC) remains a significant cause of cancer-related mortality, and early detection of precancerous polyps is crucial in reducing its impact (1-3). Although colonoscopy is the gold standard for CRC screening, studies estimate that 6–27% of polyps may be missed due to factors like operator variability and polyp characteristics (4). To address this limitation, researchers have explored artificial intelligence (AI)-driven polyp detection and classification systems, which have shown promise in increasing adenoma detection rates (ADRs). For example, a commercially available computer-aided detection (CADe) device, GI GeniusTM (Medtronic, Dublin, Ireland), has been integrated into clinical practice. In a randomized trial, the use of GI Genius increased ADR by approximately 14% compared to standard colonoscopy (5). Furthermore, clinical evaluations have reported sensitivity of 82%, specificity of 89.5%, and accuracy of 86.8%, underscoring its potential to enhance screening outcomes and reduce missed lesions, illustrating its potential impact on improving screening outcomes (6).
Recent state-of-the-art AI systems have demonstrated significant clinical benefits: one randomized trial showed that CADe-assisted colonoscopy increased ADRs from 28% to 34%—identifying subtle lesions that were often small, flat, and with unclear boundaries (7)—while another study reported that CADe assistance raised the mean number of adenomas per procedure from 1.21 to 1.56 and increased the proportion of patients with adenomas from 48.4% to 56.6% without increasing adverse events (8). Moreover, cost-effectiveness analyses suggest that integrating AI into screening colonoscopy may offer modest financial savings and reduce CRC incidence and mortality (9).
Despite these encouraging results, many current AI systems rely on specialized, high-priced hardware and are integrated into fixed endoscopic equipment—factors that limit affordability and widespread adoption. To overcome these barriers, there is growing interest in developing open-source, platform-independent solutions that can run on commonly available devices, such as smartphones (10-12) (Figure 1). Initiatives like the “Ask Doctor Smartphone!” application demonstrate the feasibility of integrating clinical decision support into handheld device (13). However, no study to date has established an open-source, real-time polyp detection framework that can operate on everyday devices while approximating realistic clinical conditions.
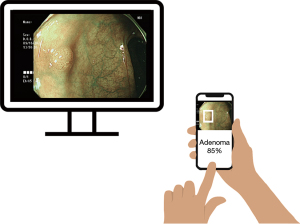
In this pilot study, we investigated the feasibility of developing an open-source, real-time polyp detection system using a versatile, platform-independent framework. By leveraging transfer learning and focusing on conditions that approximate real-world usage, we aimed to create a low-cost, flexible solution that can be deployed across various devices, including smartphones, laptops, tablets, and embedded systems. This approach seeks to democratize access to advanced diagnostic technologies, providing a customizable and accessible alternative to proprietary systems and facilitating broader adoption in diverse clinical settings.
Methods
Datasets
Dataset P1
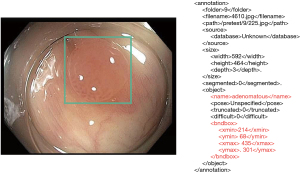
The primary dataset (P1) was obtained from the Harvard Dataverse “PolypsSet” (14), comprising 28,773 images and their corresponding Extensible Markup Language (XML) files containing information about bounding boxes and pathological classifications (adenomatous or hyperplastic) (Figure 2). Most of the images originated from sequential frames in endoscopic videos. Since the images were already randomly shuffled, no additional shuffling was performed before splitting into training and validation sets. All images were sized between 400–640 pixels in width and 350–480 pixels in height, with no additional preprocessing performed.
Datasets P2 and P2’
P2 included 130 images randomly selected from P1 to represent a variety of polyp appearances and pathologies. P2 images were normalized to maintain consistency with P1 preprocessing.
P2’ was created to simulate real-world conditions by displaying P2 images on a computer monitor and photographing them with a Galaxy S9 smartphone at approximately 50 cm distance and moderate screen brightness (~150 cd/m2). This process introduced realistic conditions such as display quality, ambient lighting, and camera-induced variations. The original P2’ data was of significantly higher resolution compared to other datasets (P1 and P2), requiring a preprocessing step to resize the images to 640×480 pixels. Additionally, to generate XML files containing bounding boxes and annotations for each P2’ image, the free website Roboflow.com was utilized (Figure 3).
Inclusion and exclusion criteria
Images were included only if they contained clearly visible, pathologically confirmed polyps (adenomatous or hyperplastic) with well-defined annotations. Images were excluded if they (I) lacked definitive pathological confirmation, (II) had unclear or missing annotations, or (III) were of poor quality due to excessive blurriness, occlusion, or lighting artifacts. Based on these criteria, 1,658 images were removed from the original P1 dataset (28,773 images), ensuring that the remaining dataset maintained high annotation accuracy and image quality.
Training and validation splits
P1 data was divided into 60% for training (n=16,273), 20% for validation (n=5,433), and 20% for testing (n=5,409). For P2 and P2’, 80% of the 130 images were used for training and 20% for validation.
Model and tools
We employed the EfficientDet Lite2 model, optimized for mobile inference, as our base architecture (15,16). Transfer learning was performed with TensorFlow Lite Model Maker with a batch size of 5 and trained for 20 epochs (17). We set the hyperparameters allowing the entire model’s weights to be updated during training (train_whole_model = true), as freezing certain layers (train_whole_model = false) led to inferior results. The training process was conducted on a consumer-grade desktop computer with an AMD Ryzen 1700 CPU, 32GB RAM, and no GPU acceleration. Due to the limitations of this setup, using a large epoch count or batch size was not feasible (Appendix 1).
spec = object_detector.EfficientDetLite2Spec()
model = object_detector.create(train_data=train_data,
model_spec=spec,
validation_data=validation_data,
epochs=20,
batch_size=5,
train_whole_model=True)
Evaluation methods and metrics
Laboratory-based evaluation
A subset of 5,409 unused P1 images (a test set) was used to evaluate models trained on P1, P2, and P2’ under controlled conditions. To assess the performance of each model, metrics such as average precision (AP) and average recall (AR) were used. AP was calculated at 50% and 75% intersection-over-union (IoU) thresholds (AP50, AP75), while AR evaluated the model’s sensitivity. Both AP and AR were also computed separately for adenomatous and hyperplastic polyps and for different polyp sizes.
Real-world evaluation
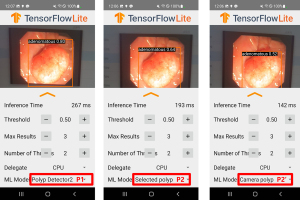
To approximate clinical usage and avoid overlap with P1, we obtained a test dataset of 100 pathologically confirmed polyp images (46 adenomas, 54 hyperplastic) from an independent source (the authors’ endoscopic videos). A Galaxy S9 smartphone running the TensorFlow Lite app then analyzed these images, which were displayed on a monitor. To simulate an endoscopy room, ambient lighting was minimized, reflecting typical dark-room conditions. The monitor brightness was set to ~150 cd/m2, and the smartphone was positioned ~50 cm away. These settings introduced realistic factors such as screen glare and camera-induced variations (Figure 4).
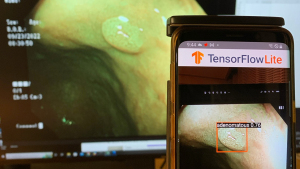
The polyp detection rate was defined as the percentage of test images where the predicted bounding box encompassed the polyp, and accuracy was the percentage of correctly classified polyps. A single experienced endoscopist (over 10,000 colonoscopies performed) verified the results. Real-time applicability was assessed by measuring inference time (Figure 5).
Statistical analysis
For comparisons of detection and classification performance between models trained on P1, P2, and P2’, Wilcoxon Rank Sum Test in the R program were employed. A P value of less than 0.05 was considered statistically significant.
Results
Laboratory-based evaluation
As shown in the table, the P1-trained model demonstrated the highest overall performance, achieving notably higher AP, AP50, and AP75 scores than either the P2- or P2’-trained models. It also maintained balanced detection capabilities for both adenomatous and hyperplastic polyps. In contrast, the P2-trained model, while substantially less accurate than P1, still outperformed P2’ under these controlled conditions. The P2’ model’s metrics consistently lagged behind those of P1 and P2 across AP and AR values, indicating weaker detection and classification abilities. Additionally, performance varied by polyp size, with all models showing diminished effectiveness for small and medium polyps compared to large ones (Table 1).
Table 1
| Metric | P1 model | P2 model | P2’ model |
|---|---|---|---|
| AP | 0.7366118 | 0.26953295 | 0.10846421 |
| AP50 | 0.9789813 | 0.43148315 | 0.19873682 |
| AP75 | 0.8823448 | 0.3190581 | 0.12356417 |
| APs (small) | 0.9 | 0 | 0 |
| APm (medium) | 0.6319911 | 0.1590814 | 0.029271524 |
| APl (large) | 0.77868646 | 0.33880654 | 0.14760345 |
| ARmax1 | 0.76531565 | 0.37641677 | 0.19987446 |
| ARmax10 | 0.7690997 | 0.4229555 | 0.26348636 |
| ARmax100 | 0.7690997 | 0.42833573 | 0.27508968 |
| ARs (small) | 0.9 | 0 | 0 |
| ARm (medium) | 0.66597825 | 0.28434783 | 0.14597826 |
| ARl (large) | 0.81041056 | 0.4960044 | 0.328849 |
| AP (adenomatous) | 0.7258867 | 0.2620216 | 0.122351006 |
| AP (hyperplastic) | 0.7473368 | 0.2770443 | 0.09457741 |
AP, average precision; AP (adenomatous), AP for adenomatous polyps; AP (hyperplastic), AP for hyperplastic polyps; AP50, AP at 50% IoU threshold; AP75, AP at 75% IoU threshold; APl, AP for large polyps; APm, AP for medium polyps; APs, AP for small polyps; AR, average recall; ARl, AR for large polyps; ARm, AR for medium polyps; ARMAX1, AR with a maximum of one detection per image; ARMAX10, AR with a maximum of 10 detections per image; ARMAX100, AR with a maximum of 100 detections per image; ARs, AR for small polyps; IoU, intersection-over-union.
Real-world smartphone evaluation
In addition to the laboratory-based evaluation, the trained models were deployed on a Galaxy S9 smartphone to assess performance under more realistic conditions. Table 2 summarizes the results of this smartphone-based test, including overall polyp detection rates, accuracy for adenomas and hyperplastic polyps, and correct classification rates. The P values indicate comparisons between the P2 and P2’ models (Table 2).
Table 2
| Metrics | P1 (%) | P2 (%) | P2’ (%) | P value (P2 vs. P2’) |
|---|---|---|---|---|
| Overall polyp detection rates | 73.20 | 32.01 | 40.21 | 0.24 |
| Accuracy for adenoma | 45.65 | 32.61 | 41.30 | 0.39 |
| Accuracy for hyperplastic polyp | 64.71 | 1.96 | 17.65 | 0.008 |
| Correct classification rates | 56.01 | 16.50 | 28.87 | 0.04 |
Under these real-world conditions, the P1-trained model maintained relatively higher performance metrics compared to the smaller datasets. Notably, the P2’ model outperformed P2 across multiple metrics, including overall polyp detection rate (40.21% vs. 32.01%) and correct classification rate (28.87% vs. 16.50%), despite the P2 model achieving better performance than P2’ in the laboratory-based evaluation. Although the differences in overall detection and classification metrics between P2 and P2’ did not always reach statistical significance (P values of 0.24 for detection rate and 0.39 for adenoma accuracy), the difference for hyperplastic polyp accuracy was statistically significant (P<0.001), and correct classification rates also reached significance (P<0.040).
The model detection on the mobile device was conducted using the smartphone’s CPU. Initially, the inference time remained within the range of 80–100 ms. However, over prolonged usage, the CPU temperature increased, resulting in a noticeable rise in inference time, which extended to 150–250 ms. Overall, the inference time per image ranged from 80 to 250 ms.
Discussion
This study demonstrates the feasibility of developing an open-source, platform-independent AI system for polyp detection using commonly available devices (18).
Our P1-trained model performed relatively well compared to smaller-scale datasets, reinforcing the established principle that larger, more diverse training sets improve model robustness and accuracy. Nonetheless, performance did not yet match top-tier commercial systems.
Compared to existing proprietary platforms, our approach is notable for its accessibility. Commercially available systems, such as GI Genius (Medtronic) and other hardware-dependent AI tools, have demonstrated significant improvements in ADRs but often require specialized equipment and can be cost-prohibitive for smaller clinics or resource-limited settings. Using smartphones and open-source frameworks could eventually lower the cost barrier, extend benefits to underserved regions, and integrate seamlessly into clinical workflows. Additionally, other open-source initiatives, such as “Ask Doctor Smartphone!”, have shown how mobile platforms can effectively deliver clinical decision support without the need for expensive hardware upgrades. Similar to the approach taken in that application, our framework aims to provide readily available AI assistance without heavy investment in specialized equipment, potentially democratizing advanced diagnostic capabilities.
However, significant challenges remain. Although P1-based training yielded moderate accuracy, it still fell short of established benchmarks. The limited performance of P2 and P2’ underscores the difficulty of training robust models with small datasets and the importance of capturing real-world variability. Introducing more data augmentation, exploring ensemble methods, or incorporating multi-modal inputs (e.g., adding non-visual clinical data) could improve performance. Moreover, better tuning of hyperparameters, selective layer unfreezing, and improved data annotation strategies may close the gap.
Real-time feedback to the endoscopist is crucial. While we achieved near real-time inference on a smartphone, integrating the model directly into the endoscopic feed for instant polyp highlighting and classification remains a work in progress. Future efforts should focus on tighter integration, where the endoscopist can adjust the scope or imaging conditions immediately based on AI prompts, potentially increasing detection and classification accuracy.
Limitations of this study include the relatively narrow variety of polyp types (only adenomas and hyperplastic polyps), reliance on a single type of consumer hardware, and challenges associated with generalizing results across different populations and endoscope models. Increasing the training dataset size, improving annotation quality, and testing on heterogeneous hardware configurations will be priorities in subsequent studies. Nevertheless, by building on prior open-source initiatives and comparing with established proprietary systems, we believe this approach can further evolve into a practical, cost-effective solution for broader clinical adoption.
Conclusions
In conclusion, this study demonstrates the feasibility of developing an open-source, smartphone-compatible AI framework for polyp detection and offers valuable insights into how training conditions influence model performance. While smaller, less diverse datasets may limit accuracy, exposing models to realistic conditions, as with P2’, can significantly improve adaptability in real-world scenarios. These findings underscore the importance of aligning training data and methodologies with actual clinical settings, as doing so can help AI-assisted colonoscopy tools become more accessible, reliable, and widely adoptable. By expanding the scope of training datasets, incorporating more advanced techniques, and considering a range of hardware configurations, future work can further enhance the effectiveness of these open-source, platform-independent solutions and ultimately improve patient outcomes.
Acknowledgments
None.
Footnote
Peer Review File: Available at https://jmai.amegroups.com/article/view/10.21037/jmai-24-310/prf
Funding: None.
Conflicts of Interest: The author has completed the ICMJE uniform disclosure form (available at https://jmai.amegroups.com/article/view/10.21037/jmai-24-310/coif). The author has no conflicts of interest to declare.
Ethical Statement: The author is accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Corley DA, Jensen CD, Marks AR, et al. Adenoma detection rate and risk of colorectal cancer and death. N Engl J Med 2014;370:1298-306. [Crossref] [PubMed]
- Siegel RL, Wagle NS, Cercek A, et al. Colorectal cancer statistics, 2023. CA Cancer J Clin 2023;73:233-54. [Crossref] [PubMed]
- Zauber AG, Winawer SJ, O'Brien MJ, et al. Colonoscopic polypectomy and long-term prevention of colorectal-cancer deaths. N Engl J Med 2012;366:687-96. [Crossref] [PubMed]
- Zhao S, Wang S, Pan P, et al. Magnitude, Risk Factors, and Factors Associated With Adenoma Miss Rate of Tandem Colonoscopy: A Systematic Review and Meta-analysis. Gastroenterology 2019;156:1661-1674.e11. [Crossref] [PubMed]
- Repici A, Badalamenti M, Maselli R, et al. Efficacy of Real-Time Computer-Aided Detection of Colorectal Neoplasia in a Randomized Trial. Gastroenterology 2020;159:512-520.e7. [Crossref] [PubMed]
- Hassan C, Balsamo G, Lorenzetti R, et al. Artificial Intelligence Allows Leaving-In-Situ Colorectal Polyps. Clin Gastroenterol Hepatol 2022;20:2505-2513.e4. [Crossref] [PubMed]
- Wang P, Liu X, Berzin TM, et al. Effect of a deep-learning computer-aided detection system on adenoma detection during colonoscopy (CADe-DB trial): a double-blind randomised study. Lancet Gastroenterol Hepatol 2020;5:343-51. [Crossref] [PubMed]
- Seager A, Sharp L, Neilson LJ, et al. Polyp detection with colonoscopy assisted by the GI Genius artificial intelligence endoscopy module compared with standard colonoscopy in routine colonoscopy practice (COLO-DETECT): a multicentre, open-label, parallel-arm, pragmatic randomised controlled trial. Lancet Gastroenterol Hepatol 2024;9:911-23. [Crossref] [PubMed]
- Areia M, Mori Y, Correale L, et al. Cost-effectiveness of artificial intelligence for screening colonoscopy: a modelling study. Lancet Digit Health 2022;4:e436-44. [Crossref] [PubMed]
- Hossain MS, Rahman MM, Syeed MM, et al. DeepPoly: Deep Learning-Based Polyps Segmentation and Classification for Autonomous Colonoscopy Examination. IEEE Access 2023;11:95889-902.
- Karmakar R, Nooshabadi S. Mobile-PolypNet: Lightweight Colon Polyp Segmentation Network for Low-Resource Settings. J Imaging 2022;8:169. [Crossref] [PubMed]
- Muneeb M, Feng SF, Henschel A. Deep learning pipeline for image classification on mobile phones. Computer Science & Information Technology 2022; [Crossref]
- Di Mitri M, Parente G, Bisanti C, et al. Ask Doctor Smartphone! An App to Help Physicians Manage Foreign Body Ingestions in Children. Diagnostics (Basel) 2023;13:3285. [Crossref] [PubMed]
- Li K, Fathan MI, Patel K, et al. Colonoscopy polyp detection and classification: Dataset creation and comparative evaluations. PLoS One 2021;16:e0255809. [Crossref] [PubMed]
- Tan M, Pang R, Le QV. EfficientDet: Scalable and Efficient Object Detection. 2020. doi:
10.1109/CVPR42600.2020.01079 .10.1109/CVPR42600.2020.01079 - AlDahoul N, Karim HA, De Castro A, et al. Localization and classification of space objects using EfficientDet detector for space situational awareness. Sci Rep 2022;12:21896. [Crossref] [PubMed]
TensorFlow Lite Model Maker - Kim YB. Polyp Detector on Android Smartphone (Github repository). Available online: https://github.com/kcchgs/polyp_detector
Cite this article as: Kim YB. Smartphone-based polyp detection: a first step towards an open-source AI framework. J Med Artif Intell 2025;8:49.


